repo_name
stringlengths 9
75
| topic
stringclasses 30
values | issue_number
int64 1
203k
| title
stringlengths 1
976
| body
stringlengths 0
254k
| state
stringclasses 2
values | created_at
stringlengths 20
20
| updated_at
stringlengths 20
20
| url
stringlengths 38
105
| labels
listlengths 0
9
| user_login
stringlengths 1
39
| comments_count
int64 0
452
|
|---|---|---|---|---|---|---|---|---|---|---|---|
DistrictDataLabs/yellowbrick
|
matplotlib
| 979
|
Visualize the results without fitting the model
|
Let's say I have to visualize a confusion matrix.
I can use yellowbrick and use the LogisticRegression and visualize like this:
https://www.scikit-yb.org/en/latest/api/classifier/confusion_matrix.html
```
from sklearn.datasets import load_iris
from sklearn.model_selection import train_test_split as tts
from sklearn.linear_model import LogisticRegression
from yellowbrick.classifier import ConfusionMatrix
iris = load_iris()
X = iris.data
y = iris.target
classes = iris.target_names
X_train, X_test, y_train, y_test = tts(X, y, test_size=0.2)
model = LogisticRegression(multi_class="auto", solver="liblinear")
iris_cm = ConfusionMatrix(
model, classes=classes,
label_encoder={0: 'setosa', 1: 'versicolor', 2: 'virginica'}
)
iris_cm.fit(X_train, y_train)
iris_cm.score(X_test, y_test)
iris_cm.show()
```
But, most of the times I use scikit-learn and I already have confusion matrix:
For example:
```
cm = np.array([[56750, 114],
[ 95, 3]])
```
Can we now simply use this result in YELLOWBRICK, give label names visualize it?
|
closed
|
2019-10-12T18:26:54Z
|
2019-10-12T18:48:32Z
|
https://github.com/DistrictDataLabs/yellowbrick/issues/979
|
[
"type: question"
] |
bhishanpdl
| 1
|
jmcnamara/XlsxWriter
|
pandas
| 170
|
Issue with DataLabels Position in Column Chart
|
I'm trying to position the data labels at the top of each column in a column chart, however doing so creates an Excel file with invalid XML that Excel won't process. I'm using XlsxWriter version 0.5.7 with Python version 2.7.6.
Here's a sample that will create such a file:
``` python
from xlsxwriter import Workbook
import random
book = Workbook('C:\\Temp\\ex.xlsx')
sheet = book.add_worksheet('Will Error')
data_sheet = book.add_worksheet('data')
year_dict = {}
year_list = [2013, 2014]
year = []
month = []
defect_rate = []
# Creates three columns of random data for each month in two years
for y in year_list:
year_dict[y] = 0
for m in range(1, 13):
year.append(y)
month.append(m)
defect_rate.append(random.randint(0, 100))
year_dict[y] += 1
data_sheet.write_column("A1", year)
data_sheet.write_column("B1", month)
data_sheet.write_column("C1", defect_rate)
chart = book.add_chart({'type': 'column'})
chart.add_series({
'values': ['data', 0, 2, year_dict[2013] - 1, 2],
'categories': ['data', 0, 1, year_dict[2013] - 1, 1],
'name': '2013',
'data_labels': {'value': True, 'position': 'top'}
})
chart.add_series({
'values': ['data', year_dict[2013], 2, year_dict[2013] + year_dict[2014] - 1, 2],
'categories': ['data', year_dict[2013], 1, year_dict[2013] + year_dict[2014] - 1, 1],
'name': '2014',
'data_labels': {'value': True, 'position': 'top'}
})
chart.set_x_axis({'name': 'Month', 'name_font': {'size': 14, 'bold': True}})
chart.set_size({'width': 800, 'height': 600})
chart.set_title({'name': "Defect Rate By Month"})
sheet.insert_chart('A1', chart)
book.close()
```
|
closed
|
2014-10-15T18:28:30Z
|
2014-10-29T02:21:50Z
|
https://github.com/jmcnamara/XlsxWriter/issues/170
|
[
"bug",
"documentation",
"ready to close"
] |
MitsuharuEishi
| 3
|
blb-ventures/strawberry-django-plus
|
graphql
| 84
|
I don't see why we need to install a new dependency django-choices-field
|
https://github.com/blb-ventures/strawberry-django-plus/blob/9f06e1169f6ce696a9439bec52abd546ef380b29/strawberry_django_plus/types.py#L208
Hey there, I'm currently trying the lib here, I'm enjoying it so far, hopefully it gets merged with the main lib soon.
Regarding the above, can't you just do something like the following
```
isinstance(field, CharField) and Model._meta.get_field('<field_name>').choices
isinstance(field, IntegerField) and Model._meta.get_field('<field_name>').choices
```
Shouldn't that be enough? Assuming you're able to get access to to the model there.
|
open
|
2022-07-17T02:00:16Z
|
2022-07-19T22:18:36Z
|
https://github.com/blb-ventures/strawberry-django-plus/issues/84
|
[
"question"
] |
mhdismail
| 5
|
marshmallow-code/apispec
|
rest-api
| 176
|
Can't use apispec tornado plugin icw complex paths
|
I'm trying to use apispec icw the 'apispec.ext.tornado' and 'apispec.ext.marshmallow' plugins.
Only i'm getting the following error:
```
Traceback (most recent call last):
File "D:\JetBrains\PyCharm 2017.2.4\helpers\pydev\pydevd.py", line 1599, in <module>
globals = debugger.run(setup['file'], None, None, is_module)
File "D:\JetBrains\PyCharm 2017.2.4\helpers\pydev\pydevd.py", line 1026, in run
pydev_imports.execfile(file, globals, locals) # execute the script
File "D:\myproj/start.py", line 7, in <module>
main()
File "D:\myproj\medusa\__main__.py", line 2104, in main
application.start(sys.argv[1:])
File "D:\myproj\medusa\__main__.py", line 347, in start
self.web_server = AppWebServer(self.web_options)
File "D:\myproj\medusa\server\core.py", line 230, in __init__
spec.add_path(urlspec=urlspec)
File "D:\Python27\lib\site-packages\apispec\core.py", line 211, in add_path
raise APISpecError('Path template is not specified')
apispec.exceptions.APISpecError: Path template is not specified
```
It seems to be happening because matcher._path is None.
https://github.com/marshmallow-code/apispec/blob/dev/apispec/ext/tornado.py#L95
`urlspec.matcher._path` returns None, because of this:
https://github.com/tornadoweb/tornado/blob/master/tornado/routing.py#L571
And my route looks like this:
`'/api/v2/series/(?P<series_slug>\\w+)/episode(?:(?:(?:(?:/(?P<episode_slug>[\\w-]+))|/?)(?:(?:/(?P<path_param>\\w+))|/?))|/?)/?$'`
So because the tornado plugin uses the matcher._path, it's can't translate to an OpenApi compliant path.
Is there anything I can do about it?
|
closed
|
2017-12-11T19:41:43Z
|
2018-11-03T14:33:19Z
|
https://github.com/marshmallow-code/apispec/issues/176
|
[] |
p0psicles
| 2
|
ray-project/ray
|
python
| 51,642
|
[core] Unify `CoreWorker::Exit` and `CoreWorker::Shutdown`
|
### Description
See https://github.com/ray-project/ray/pull/51582#discussion_r2010500080 for more details.
### Use case
_No response_
|
open
|
2025-03-24T16:52:35Z
|
2025-03-24T16:52:44Z
|
https://github.com/ray-project/ray/issues/51642
|
[
"enhancement",
"core"
] |
kevin85421
| 0
|
FlareSolverr/FlareSolverr
|
api
| 1,223
|
[yggtorrent] (testing) Exception (yggtorrent): FlareSolverr was unable to process the request, please check FlareSolverr logs. Message: Error: Error solving the challenge. Timeout after 55.0 seconds.: FlareSolverr was unable to process the request, please check FlareSolverr logs. Message: Error: Error solving the challenge. Timeout after 55.0 seconds.
|
### Have you checked our README?
- [X] I have checked the README
### Have you followed our Troubleshooting?
- [X] I have followed your Troubleshooting
### Is there already an issue for your problem?
- [X] I have checked older issues, open and closed
### Have you checked the discussions?
- [X] I have read the Discussions
### Environment
```markdown
- FlareSolverr version:3.3.19
- Last working FlareSolverr version:3.3.19
- Operating system:debian
- Are you using Docker: no
- FlareSolverr User-Agent (see log traces or / endpoint):
- Are you using a VPN: no
- Are you using a Proxy: no
- Are you using Captcha Solver: no
- If using captcha solver, which one:
- URL to test this issue:
```
### Description
"Usual" issue with cloudflare breaking up access to yggtorrent rather regularly.
Jackett updated to v0.22.188
### Logged Error Messages
```text
Jackett.Common.IndexerException: Exception (yggtorrent): FlareSolverr was unable to process the request, please check FlareSolverr logs. Message: Error: Error solving the challenge. Timeout after 55.0 seconds.
[v0.22.188.0] Jackett.Common.IndexerException: Exception (yggtorrent): FlareSolverr was unable to process the request, please check FlareSolverr logs. Message: Error: Error solving the challenge. Timeout after 55.0 seconds.
---> FlareSolverrSharp.Exceptions.FlareSolverrException: FlareSolverr was unable to process the request, please check FlareSolverr logs. Message: Error: Error solving the challenge. Timeout after 55.0 seconds.
at FlareSolverrSharp.Solvers.FlareSolverr.<>c__DisplayClass12_0.<<SendFlareSolverrRequest>b__0>d.MoveNext()
--- End of stack trace from previous location ---
at FlareSolverrSharp.Utilities.SemaphoreLocker.LockAsync[T](Func`1 worker)
at FlareSolverrSharp.Solvers.FlareSolverr.SendFlareSolverrRequest(HttpContent flareSolverrRequest)
at FlareSolverrSharp.Solvers.FlareSolverr.Solve(HttpRequestMessage request, String sessionId)
at FlareSolverrSharp.ClearanceHandler.SendAsync(HttpRequestMessage request, CancellationToken cancellationToken)
at System.Net.Http.HttpClient.<SendAsync>g__Core|83_0(HttpRequestMessage request, HttpCompletionOption completionOption, CancellationTokenSource cts, Boolean disposeCts, CancellationTokenSource pendingRequestsCts, CancellationToken originalCancellationToken)
at Jackett.Common.Utils.Clients.HttpWebClient2.Run(WebRequest webRequest) in ./Jackett.Common/Utils/Clients/HttpWebClient2.cs:line 180
at Jackett.Common.Utils.Clients.WebClient.GetResultAsync(WebRequest request) in ./Jackett.Common/Utils/Clients/WebClient.cs:line 186
at Jackett.Common.Indexers.BaseWebIndexer.RequestWithCookiesAsync(String url, String cookieOverride, RequestType method, String referer, IEnumerable`1 data, Dictionary`2 headers, String rawbody, Nullable`1 emulateBrowser) in ./Jackett.Common/Indexers/BaseIndexer.cs:line 598
at Jackett.Common.Indexers.CardigannIndexer.PerformQuery(TorznabQuery query) in ./Jackett.Common/Indexers/CardigannIndexer.cs:line 1532
at Jackett.Common.Indexers.BaseIndexer.ResultsForQuery(TorznabQuery query, Boolean isMetaIndexer) in ./Jackett.Common/Indexers/BaseIndexer.cs:line 366
--- End of inner exception stack trace ---
at Jackett.Common.Indexers.BaseIndexer.ResultsForQuery(TorznabQuery query, Boolean isMetaIndexer) in ./Jackett.Common/Indexers/BaseIndexer.cs:line 387
at Jackett.Common.Indexers.BaseWebIndexer.ResultsForQuery(TorznabQuery query, Boolean isMetaIndexer) in ./Jackett.Common/Indexers/BaseIndexer.cs:line 778
at Jackett.Common.Services.IndexerManagerService.TestIndexer(String name) in ./Jackett.Common/Services/IndexerManagerService.cs:line 323
at Jackett.Server.Controllers.IndexerApiController.Test() in ./Jackett.Server/Controllers/IndexerApiController.cs:line 132
at Microsoft.AspNetCore.Mvc.Infrastructure.ActionMethodExecutor.TaskOfIActionResultExecutor.Execute(ActionContext actionContext, IActionResultTypeMapper mapper, ObjectMethodExecutor executor, Object controller, Object[] arguments)
at Microsoft.AspNetCore.Mvc.Infrastructure.ControllerActionInvoker.<InvokeActionMethodAsync>g__Awaited|12_0(ControllerActionInvoker invoker, ValueTask`1 actionResultValueTask)
at Microsoft.AspNetCore.Mvc.Infrastructure.ControllerActionInvoker.<InvokeNextActionFilterAsync>g__Awaited|10_0(ControllerActionInvoker invoker, Task lastTask, State next, Scope scope, Object state, Boolean isCompleted)
at Microsoft.AspNetCore.Mvc.Infrastructure.ControllerActionInvoker.Rethrow(ActionExecutedContextSealed context)
at Microsoft.AspNetCore.Mvc.Infrastructure.ControllerActionInvoker.Next(State& next, Scope& scope, Object& state, Boolean& isCompleted)
at Microsoft.AspNetCore.Mvc.Infrastructure.ControllerActionInvoker.<InvokeInnerFilterAsync>g__Awaited|13_0(ControllerActionInvoker invoker, Task lastTask, State next, Scope scope, Object state, Boolean isCompleted)
at Microsoft.AspNetCore.Mvc.Infrastructure.ResourceInvoker.<InvokeFilterPipelineAsync>g__Awaited|20_0(ResourceInvoker invoker, Task lastTask, State next, Scope scope, Object state, Boolean isCompleted)
at Microsoft.AspNetCore.Mvc.Infrastructure.ResourceInvoker.<InvokeAsync>g__Awaited|17_0(ResourceInvoker invoker, Task task, IDisposable scope)
at Microsoft.AspNetCore.Mvc.Infrastructure.ResourceInvoker.<InvokeAsync>g__Awaited|17_0(ResourceInvoker invoker, Task task, IDisposable scope)
at Microsoft.AspNetCore.Authentication.AuthenticationMiddleware.Invoke(HttpContext context)
at Microsoft.AspNetCore.Builder.Extensions.UsePathBaseMiddleware.InvokeCore(HttpContext context, PathString matchedPath, PathString remainingPath)
at Jackett.Server.Middleware.CustomExceptionHandler.Invoke(HttpContext httpContext) in ./Jackett.Server/Middleware/CustomExceptionHandler.cs:line 26
```
### Screenshots
_No response_
|
closed
|
2024-06-21T09:21:23Z
|
2024-06-21T09:26:08Z
|
https://github.com/FlareSolverr/FlareSolverr/issues/1223
|
[
"duplicate"
] |
eejag
| 1
|
python-gino/gino
|
sqlalchemy
| 351
|
Release GINO 0.7.6 and 0.8
|
I'm planning to release GINO 0.8 (1.0-rc) from current master, and close `v0.6.x` branch and support. @wwwjfy anything to add please?
|
closed
|
2018-09-29T03:28:29Z
|
2018-10-17T07:41:56Z
|
https://github.com/python-gino/gino/issues/351
|
[
"task"
] |
fantix
| 11
|
coqui-ai/TTS
|
deep-learning
| 3,840
|
[Bug] can not load the checkpoints when do fine-tune with XTTS_v2
|
### Describe the bug
Hello, community and @eginhard,
For the XTTS fine-tuning, I manually downloaded `dvae.pth, mel_stats.pth, model.pth` and `vocab.json` in `train_gpt_xtts.py`.
Further, below is the command line for fine-tuning XTTS_v2.
```
CUDA_VISIBLE_DEVICES="0" python recipes/mshop/xtts_v2/train_gpt_xtts.py \
--restore_path /home/ec2-user/SageMaker/workspace/TTS/XTTS/xtts_v2/model.pth
```
where the model.pth is derived from `tts = TTS("tts_models/multilingual/multi-dataset/xtts_v2").to(device)`
I got the error as below:
```
| > Layer missing in the checkpoint: dvae.decoder.3.net.4.weight
| > Layer missing in the checkpoint: dvae.decoder.3.net.4.bias
| > Layer missing in the checkpoint: dvae.decoder.4.0.conv.weight
| > Layer missing in the checkpoint: dvae.decoder.4.0.conv.bias
| > Layer missing in the checkpoint: dvae.decoder.5.0.conv.weight
| > Layer missing in the checkpoint: dvae.decoder.5.0.conv.bias
| > Layer missing in the checkpoint: dvae.decoder.6.weight
| > Layer missing in the checkpoint: dvae.decoder.6.bias
| > Layer missing in the checkpoint: dvae.codebook.embed
| > Layer missing in the checkpoint: dvae.codebook.cluster_size
| > Layer missing in the checkpoint: dvae.codebook.embed_avg
| > Layer missing in the checkpoint: torch_mel_spectrogram_dvae.mel_stft.spectrogram.window
| > Layer missing in the checkpoint: torch_mel_spectrogram_dvae.mel_stft.mel_scale.fb
| > 0 / 1023 layers are restored.
> Model restored from step 10000000
Traceback (most recent call last):
File "/home/ec2-user/SageMaker/workspace/TTS/XTTS/recipes/mshop/xtts_v2/train_gpt_xtts.py", line 202, in <module>
main()
File "/home/ec2-user/SageMaker/workspace/TTS/XTTS/recipes/mshop/xtts_v2/train_gpt_xtts.py", line 186, in main
trainer = Trainer(
File "/home/ec2-user/anaconda3/envs/JupyterSystemEnv/lib/python3.10/site-packages/trainer/trainer.py", line 558, in __init__
(self.model, self.optimizer, self.scaler, self.restore_step, self.restore_epoch) = self.restore_model(
File "/home/ec2-user/anaconda3/envs/JupyterSystemEnv/lib/python3.10/site-packages/trainer/trainer.py", line 862, in restore_model
restore_epoch = checkpoint["epoch"]
KeyError: 'epoch'
```
As a result,
1) I put the pretrained weights of dvae, hifigan (mel_stats.pth), model.pth in DVAE_CHECKPOINT, MEL_NORM_FILE, TOKENIZER_FILE, and XTTS_CHECKPOINT, but it seems like not working
2) When I check the XTTS_v2 checkpoints with torch.load() and do find 'epoch', there is no epoch in checkpoints.
### To Reproduce
```
CUDA_VISIBLE_DEVICES="0" python recipes/mshop/xtts_v2/train_gpt_xtts.py \
--restore_path /home/ec2-user/SageMaker/workspace/TTS/XTTS/xtts_v2/model.pth
```
### Expected behavior
fine-tuning with own dataset
### Logs
```shell
| > Layer missing in the checkpoint: xtts.gpt.gpt.h.28.ln_2.bias
| > Layer missing in the checkpoint: xtts.gpt.gpt.h.28.mlp.c_fc.weight
| > Layer missing in the checkpoint: xtts.gpt.gpt.h.28.mlp.c_fc.bias
| > Layer missing in the checkpoint: xtts.gpt.gpt.h.28.mlp.c_proj.weight
| > Layer missing in the checkpoint: xtts.gpt.gpt.h.28.mlp.c_proj.bias
| > Layer missing in the checkpoint: xtts.gpt.gpt.h.29.ln_1.weight
| > Layer missing in the checkpoint: xtts.gpt.gpt.h.29.ln_1.bias
| > Layer missing in the checkpoint: xtts.gpt.gpt.h.29.attn.c_attn.weight
| > Layer missing in the checkpoint: xtts.gpt.gpt.h.29.attn.c_attn.bias
| > Layer missing in the checkpoint: xtts.gpt.gpt.h.29.attn.c_proj.weight
| > Layer missing in the checkpoint: xtts.gpt.gpt.h.29.attn.c_proj.bias
| > Layer missing in the checkpoint: xtts.gpt.gpt.h.29.ln_2.weight
| > Layer missing in the checkpoint: xtts.gpt.gpt.h.29.ln_2.bias
| > Layer missing in the checkpoint: xtts.gpt.gpt.h.29.mlp.c_fc.weight
| > Layer missing in the checkpoint: xtts.gpt.gpt.h.29.mlp.c_fc.bias
| > Layer missing in the checkpoint: xtts.gpt.gpt.h.29.mlp.c_proj.weight
| > Layer missing in the checkpoint: xtts.gpt.gpt.h.29.mlp.c_proj.bias
| > Layer missing in the checkpoint: xtts.gpt.gpt.ln_f.weight
| > Layer missing in the checkpoint: xtts.gpt.gpt.ln_f.bias
| > Layer missing in the checkpoint: xtts.gpt.mel_pos_embedding.emb.weight
| > Layer missing in the checkpoint: xtts.gpt.text_pos_embedding.emb.weight
| > Layer missing in the checkpoint: xtts.gpt.final_norm.weight
| > Layer missing in the checkpoint: xtts.gpt.final_norm.bias
| > Layer missing in the checkpoint: xtts.gpt.text_head.weight
| > Layer missing in the checkpoint: xtts.gpt.text_head.bias
| > Layer missing in the checkpoint: xtts.gpt.mel_head.weight
| > Layer missing in the checkpoint: xtts.gpt.mel_head.bias
| > Layer missing in the checkpoint: xtts.gpt.conditioning_perceiver.latents
| > Layer missing in the checkpoint: xtts.gpt.conditioning_perceiver.layers.0.0.to_q.weight
| > Layer missing in the checkpoint: xtts.gpt.conditioning_perceiver.layers.0.0.to_kv.weight
| > Layer missing in the checkpoint: xtts.gpt.conditioning_perceiver.layers.0.0.to_out.weight
| > Layer missing in the checkpoint: xtts.gpt.conditioning_perceiver.layers.0.1.0.weight
| > Layer missing in the checkpoint: xtts.gpt.conditioning_perceiver.layers.0.1.0.bias
| > Layer missing in the checkpoint: xtts.gpt.conditioning_perceiver.layers.0.1.2.weight
| > Layer missing in the checkpoint: xtts.gpt.conditioning_perceiver.layers.0.1.2.bias
| > Layer missing in the checkpoint: xtts.gpt.conditioning_perceiver.layers.1.0.to_q.weight
| > Layer missing in the checkpoint: xtts.gpt.conditioning_perceiver.layers.1.0.to_kv.weight
| > Layer missing in the checkpoint: xtts.gpt.conditioning_perceiver.layers.1.0.to_out.weight
| > Layer missing in the checkpoint: xtts.gpt.conditioning_perceiver.layers.1.1.0.weight
| > Layer missing in the checkpoint: xtts.gpt.conditioning_perceiver.layers.1.1.0.bias
| > Layer missing in the checkpoint: xtts.gpt.conditioning_perceiver.layers.1.1.2.weight
| > Layer missing in the checkpoint: xtts.gpt.conditioning_perceiver.layers.1.1.2.bias
| > Layer missing in the checkpoint: xtts.gpt.conditioning_perceiver.norm.gamma
| > Layer missing in the checkpoint: torch_mel_spectrogram_style_encoder.mel_stft.spectrogram.window
| > Layer missing in the checkpoint: torch_mel_spectrogram_style_encoder.mel_stft.mel_scale.fb
| > Layer missing in the checkpoint: dvae.discrete_loss.accumulator_index
| > Layer missing in the checkpoint: dvae.discrete_loss.accumulator_filled
| > Layer missing in the checkpoint: dvae.discrete_loss.accumulator
| > Layer missing in the checkpoint: dvae.encoder.0.0.weight
| > Layer missing in the checkpoint: dvae.encoder.0.0.bias
| > Layer missing in the checkpoint: dvae.encoder.1.0.weight
| > Layer missing in the checkpoint: dvae.encoder.1.0.bias
| > Layer missing in the checkpoint: dvae.encoder.2.net.0.weight
| > Layer missing in the checkpoint: dvae.encoder.2.net.0.bias
| > Layer missing in the checkpoint: dvae.encoder.2.net.2.weight
| > Layer missing in the checkpoint: dvae.encoder.2.net.2.bias
| > Layer missing in the checkpoint: dvae.encoder.2.net.4.weight
| > Layer missing in the checkpoint: dvae.encoder.2.net.4.bias
| > Layer missing in the checkpoint: dvae.encoder.3.net.0.weight
| > Layer missing in the checkpoint: dvae.encoder.3.net.0.bias
| > Layer missing in the checkpoint: dvae.encoder.3.net.2.weight
| > Layer missing in the checkpoint: dvae.encoder.3.net.2.bias
| > Layer missing in the checkpoint: dvae.encoder.3.net.4.weight
| > Layer missing in the checkpoint: dvae.encoder.3.net.4.bias
| > Layer missing in the checkpoint: dvae.encoder.4.net.0.weight
| > Layer missing in the checkpoint: dvae.encoder.4.net.0.bias
| > Layer missing in the checkpoint: dvae.encoder.4.net.2.weight
| > Layer missing in the checkpoint: dvae.encoder.4.net.2.bias
| > Layer missing in the checkpoint: dvae.encoder.4.net.4.weight
| > Layer missing in the checkpoint: dvae.encoder.4.net.4.bias
| > Layer missing in the checkpoint: dvae.encoder.5.weight
| > Layer missing in the checkpoint: dvae.encoder.5.bias
| > Layer missing in the checkpoint: dvae.decoder.0.weight
| > Layer missing in the checkpoint: dvae.decoder.0.bias
| > Layer missing in the checkpoint: dvae.decoder.1.net.0.weight
| > Layer missing in the checkpoint: dvae.decoder.1.net.0.bias
| > Layer missing in the checkpoint: dvae.decoder.1.net.2.weight
| > Layer missing in the checkpoint: dvae.decoder.1.net.2.bias
| > Layer missing in the checkpoint: dvae.decoder.1.net.4.weight
| > Layer missing in the checkpoint: dvae.decoder.1.net.4.bias
| > Layer missing in the checkpoint: dvae.decoder.2.net.0.weight
| > Layer missing in the checkpoint: dvae.decoder.2.net.0.bias
| > Layer missing in the checkpoint: dvae.decoder.2.net.2.weight
| > Layer missing in the checkpoint: dvae.decoder.2.net.2.bias
| > Layer missing in the checkpoint: dvae.decoder.2.net.4.weight
| > Layer missing in the checkpoint: dvae.decoder.2.net.4.bias
| > Layer missing in the checkpoint: dvae.decoder.3.net.0.weight
| > Layer missing in the checkpoint: dvae.decoder.3.net.0.bias
| > Layer missing in the checkpoint: dvae.decoder.3.net.2.weight
| > Layer missing in the checkpoint: dvae.decoder.3.net.2.bias
| > Layer missing in the checkpoint: dvae.decoder.3.net.4.weight
| > Layer missing in the checkpoint: dvae.decoder.3.net.4.bias
| > Layer missing in the checkpoint: dvae.decoder.4.0.conv.weight
| > Layer missing in the checkpoint: dvae.decoder.4.0.conv.bias
| > Layer missing in the checkpoint: dvae.decoder.5.0.conv.weight
| > Layer missing in the checkpoint: dvae.decoder.5.0.conv.bias
| > Layer missing in the checkpoint: dvae.decoder.6.weight
| > Layer missing in the checkpoint: dvae.decoder.6.bias
| > Layer missing in the checkpoint: dvae.codebook.embed
| > Layer missing in the checkpoint: dvae.codebook.cluster_size
| > Layer missing in the checkpoint: dvae.codebook.embed_avg
| > Layer missing in the checkpoint: torch_mel_spectrogram_dvae.mel_stft.spectrogram.window
| > Layer missing in the checkpoint: torch_mel_spectrogram_dvae.mel_stft.mel_scale.fb
| > 0 / 1023 layers are restored.
> Model restored from step 10000000
Traceback (most recent call last):
File "/home/ec2-user/SageMaker/workspace/TTS/XTTS/recipes/mshop/xtts_v2/train_gpt_xtts.py", line 202, in <module>
main()
File "/home/ec2-user/SageMaker/workspace/TTS/XTTS/recipes/mshop/xtts_v2/train_gpt_xtts.py", line 186, in main
trainer = Trainer(
File "/home/ec2-user/anaconda3/envs/JupyterSystemEnv/lib/python3.10/site-packages/trainer/trainer.py", line 558, in __init__
(self.model, self.optimizer, self.scaler, self.restore_step, self.restore_epoch) = self.restore_model(
File "/home/ec2-user/anaconda3/envs/JupyterSystemEnv/lib/python3.10/site-packages/trainer/trainer.py", line 862, in restore_model
restore_epoch = checkpoint["epoch"]
KeyError: 'epoch'
```
### Environment
```shell
{
"CUDA": {
"GPU": [
"Tesla V100-SXM2-32GB",
"Tesla V100-SXM2-32GB",
"Tesla V100-SXM2-32GB",
"Tesla V100-SXM2-32GB",
"Tesla V100-SXM2-32GB",
"Tesla V100-SXM2-32GB",
"Tesla V100-SXM2-32GB",
"Tesla V100-SXM2-32GB"
],
"available": true,
"version": "12.1"
},
"Packages": {
"PyTorch_debug": false,
"PyTorch_version": "2.3.1+cu121",
"TTS": "0.22.0",
"numpy": "1.22.0"
},
"System": {
"OS": "Linux",
"architecture": [
"64bit",
"ELF"
],
"processor": "x86_64",
"python": "3.10.8",
"version": "#1 SMP Tue May 21 16:52:24 UTC 2024"
}
}
```
### Additional context
...
|
closed
|
2024-07-29T14:10:11Z
|
2024-07-29T18:46:28Z
|
https://github.com/coqui-ai/TTS/issues/3840
|
[
"bug"
] |
kaen2891
| 4
|
jina-ai/serve
|
fastapi
| 5,613
|
Error reporting when DNS exists, but route is not valid
|
When an external Executor/Flow is down, but there is no DNS error (e.g. because it is behind an API gateway), then the error reporting does not show which Executor is the failing one.
**Reproduce:**
```python
from jina import Flow
from docarray import DocumentArray, Document
f = Flow().add(host='https://blah.wolf.jina.ai/', external=True) # this Flow does not exist, but no DNS issue
with f:
f.post(inputs=Document, on='/foo')
```
On the client:
```text
jina.excepts.BadServerFlow: gRPC error: StatusCode.UNKNOWN Unexpected <class 'grpc.aio._call.AioRpcError'>: <AioRpcError of RPC that terminated with:
status = StatusCode.NOT_FOUND
details = "no Route matched with those values"
debug_error_string = "{"created":"@1674229068.908652727","description":"Error received from peer ipv4:35.169.210.186:443","file":"src/core/lib/surface/call.cc","file_line":952,"grpc_message":"no Route matched with those values","grpc_status":5}"
>
```
On the gateway:
```
ERROR gateway/rep-0/GatewayRuntime@91339 Error while
getting responses from deployments: <AioRpcError of
RPC that terminated with:
status = StatusCode.NOT_FOUND
details = "no Route matched with those
values"
debug_error_string =
"{"created":"@1674229068.908652727","description":"E…
received from peer
ipv4:35.169.210.186:443","file":"src/core/lib/surfac…
Route matched with those values","grpc_status":5}"
>
```
Reference: https://jinaai.slack.com/archives/C018F60RBL5/p1674205594827639
|
closed
|
2023-01-20T12:05:32Z
|
2023-01-24T11:40:54Z
|
https://github.com/jina-ai/serve/issues/5613
|
[] |
JohannesMessner
| 0
|
tensorflow/tensor2tensor
|
deep-learning
| 1,640
|
mismatch of tensor2tensor.layers.common_audio.compute_mel_filterbank_features with tensorflow audio_ops.mfcc
|
### Description
...
### Environment information
```
OS: Ubuntu 18.04
$ pip freeze | grep tensor
tensorflow: 1.14.0
$ python -V
3.6.8
```
### For bugs: reproduction and error logs
```
# Steps to reproduce:
import tensorflow as tf
tf.enable_eager_execution()
from tensorflow.contrib.framework.python.ops import audio_ops
import numpy as np
import matplotlib.pyplot as plt
from tensor2tensor.layers.common_audio import compute_mel_filterbank_features
sample_rate = 16000
desired_samples = 16000
nyquist = sample_rate // 2
num_sinusoids = 5
frame_length = 512
frame_step = 320
fft_length = 512
# Sinusoid sweep from 1 Hz to nyquist.
frequencies = tf.lin_space(1.0, nyquist - 1, num_sinusoids)
# Generate the sinusoids.
signal = tf.reduce_sum(tf.math.sin(2.0 * np.pi * tf.range(desired_samples, dtype=tf.float32)[tf.newaxis, :] * frequencies[:, tf.newaxis] / sample_rate), 0)
# Add some white noise for fun.
signal += tf.random_normal([desired_samples]) * 0.1
print(signal.get_shape())
num_mfccs = 26
lower_edge_hertz=20
upper_edge_hertz=4000.0
log_noise_floor=1e-4
num_mel_bins = 40
sample_rate=16000
spectrogram = tf.squeeze(audio_ops.audio_spectrogram(signal[:, tf.newaxis],
window_size=frame_length,
stride=frame_step,
magnitude_squared=False), 0)
audio_ops_mfccs = audio_ops.mfcc(tf.expand_dims(spectrogram, 0), \
sample_rate=sample_rate, \
lower_frequency_limit=lower_edge_hertz, \
upper_frequency_limit=upper_edge_hertz, \
filterbank_channel_count=num_mel_bins, \
dct_coefficient_count=num_mfccs)
audio_ops_mfccs= tf.squeeze(audio_ops_mfccs,0 )
signal_mfccs = compute_mel_filterbank_features(
signal,
sample_rate=sample_rate,
frame_length=frame_length,
frame_step=frame_step,
lower_edge_hertz=lower_edge_hertz,
upper_edge_hertz=upper_edge_hertz,
num_mel_bins=num_mfccs,
apply_mask=False)
signal_mfccs =tf.signal.mfccs_from_log_mel_spectrograms(signal_mfccs)
np.testing.assert_allclose(signal_mfccs, audio_ops_mfccs, rtol=1e-4, atol=1e-4);
```
```
# Error logs:
mismatch in the results of both the apis, as per the tensorflow [issue-11339](https://github.com/tensorflow/tensorflow/issues/11339#issuecomment-345741527), audio ops is just a FusedOP for mobile computation the algorithm is still same and also the float precision is different. the way tensor2tensor computes the mel spectrograms doesn't matches the results for tf audio ops.
```
|
open
|
2019-07-24T09:07:23Z
|
2019-07-24T09:07:23Z
|
https://github.com/tensorflow/tensor2tensor/issues/1640
|
[] |
cahuja1992
| 0
|
custom-components/pyscript
|
jupyter
| 420
|
Enhancement request: wildcards in @state_trigger
|
Can `@state_trigger` be changed to support wildcards?
I am wanting to do:
````
@state_trigger('sensor.ble_temperature_*')
````
|
closed
|
2022-12-31T17:36:05Z
|
2023-01-01T04:21:16Z
|
https://github.com/custom-components/pyscript/issues/420
|
[] |
fovea1959
| 3
|
PrefectHQ/prefect
|
automation
| 16,917
|
Prefect UI only shows blank white screen
|
> Hey! I'm getting the same result by running prefect inside a uv venv on linux, commands used:
> ```
> uv venv --python 3.12 && source .venv/bin/activate
> uv pip install -U prefect
> prefect server start
> ```
>
> Visiting localhost or 127.0.0.1 gives the same result, /docs works as intended. Screenshot with error:
>
> 
_Originally posted by @HRKings in [#10452](https://github.com/PrefectHQ/prefect/issues/10452#issuecomment-2625606594)_
|
open
|
2025-01-30T21:58:40Z
|
2025-01-30T21:58:40Z
|
https://github.com/PrefectHQ/prefect/issues/16917
|
[] |
aaazzam
| 0
|
tflearn/tflearn
|
tensorflow
| 215
|
validation_set doesn't seem to run through Image Augmentation
|
I'm trying to use validation_set=0.1 with the below tflearn image augmentation pipeline, but I get the following error which makes it seem like tflearn is trying to run the metric on the original image (128,128,3) instead of an augmented one (112,112,3):
Cannot feed value of shape (128, 128, 128, 3) for Tensor u'InputData/X:0', which has shape '(?, 112, 112, 3)'
``` python
img_aug = ImageAugmentation()
img_aug.add_random_crop((112,112))
net = input_data(shape=[None, 112, 112, 3], data_augmentation=img_aug)
...
...
net = regression(net, optimizer='adam', loss='mean_square', learning_rate=0.001)
model = tflearn.DNN(net, tensorboard_verbose=0)
model.fit(X, latent, n_epoch=50, shuffle=True, show_metric=True,
batch_size=128, validation_set=0.1)
```
|
open
|
2016-07-21T11:33:56Z
|
2016-07-21T17:01:23Z
|
https://github.com/tflearn/tflearn/issues/215
|
[] |
tetmin
| 3
|
iMerica/dj-rest-auth
|
rest-api
| 67
|
Docs Improvement: Register users from admin panel in a compatible way.
|
Hi, I went through the documentation for this library. But didn't find an answer for my question.
So I implemented dj-rest-auth and everything is smooth AF.
Thank you so much for this amazing library.
So a user can register via my web app..but I want to be able to add users from my admin application. How can I go about doing that. I have User registered to admin app, but I guess that's not enough.
What are the next steps I need to follow.
|
open
|
2020-05-12T05:53:04Z
|
2022-03-30T10:08:55Z
|
https://github.com/iMerica/dj-rest-auth/issues/67
|
[
"enhancement"
] |
Goutam192002
| 9
|
milesmcc/shynet
|
django
| 329
|
Docker-less troubles
|
Edit: updated instructions to fix my own problem. And help anyone who finds this.
As discussed in #9, I've been trying to get shynet up and running without docker in order to submit a PR with documentation. I'm using RHEL but once done the instructions can easily be translated into Debian or any other Linux flavor.
I'm running into issues. Here's what I've done so far:
1. ```sudo dnf install -y python3 python3-pip git gcc```
2. ```curl -sSL https://install.python-poetry.org | python3 -```
3. ```git clone https://github.com/milesmcc/shynet.git```
4. ```cd shynet```
5. ```npm install```
6. ```poetry run pip install "Cython<3.0" "pyyaml==5.4.1" "django-allauth==0.45.0" --no-build-isolation```
7. ```poetry install```
8. set up the ```.env``` file with your ```db```, your domain in ```allowed_hosts``` and ```csrf_trusted_origins```, and ```port``` (matching your vhost). I also did ```django_secret_key``` and ```time_zone```.
9. set up a corresponding vhost or Caddy site block
10. ```poetry run python manage.py migrate```
11. ```poetry run python manage.py collectstatic```
12. ```python manage.py compilemessages```
~~Here's the output:~~ The updated instructions shouldn't cause this error.
```File "shynet/shynet/manage.py", line 21, in <module>
main()
File "shynet/shynet/manage.py", line 17, in main
execute_from_command_line(sys.argv)
File "/root/.cache/pypoetry/virtualenvs/shynet-p4mndYDs-py3.9/lib/python3.9/site-packages/django/core/management/__init__.py", line 446, in execute_from_command_line
utility.execute()
File "/root/.cache/pypoetry/virtualenvs/shynet-p4mndYDs-py3.9/lib/python3.9/site-packages/django/core/management/__init__.py", line 420, in execute
django.setup()
File "/root/.cache/pypoetry/virtualenvs/shynet-p4mndYDs-py3.9/lib/python3.9/site-packages/django/__init__.py", line 24, in setup
apps.populate(settings.INSTALLED_APPS)
File "/root/.cache/pypoetry/virtualenvs/shynet-p4mndYDs-py3.9/lib/python3.9/site-packages/django/apps/registry.py", line 91, in populate
app_config = AppConfig.create(entry)
File "/root/.cache/pypoetry/virtualenvs/shynet-p4mndYDs-py3.9/lib/python3.9/site-packages/django/apps/config.py", line 193, in create
import_module(entry)
File "/usr/lib64/python3.9/importlib/__init__.py", line 127, in import_module
return _bootstrap._gcd_import(name[level:], package, level)
File "<frozen importlib._bootstrap>", line 1030, in _gcd_import
File "<frozen importlib._bootstrap>", line 1007, in _find_and_load
File "<frozen importlib._bootstrap>", line 984, in _find_and_load_unlocked
ModuleNotFoundError: No module named 'allauth'
```
``` - Installing django-allauth (0.45.0): Failed
ChefBuildError
Backend subprocess exited when trying to invoke get_requires_for_build_wheel
Traceback (most recent call last):
File "/root/.local/share/pypoetry/venv/lib64/python3.9/site-packages/pyproject_hooks/_in_process/_in_process.py", line 373, in <module>
main()
File "/root/.local/share/pypoetry/venv/lib64/python3.9/site-packages/pyproject_hooks/_in_process/_in_process.py", line 357, in main
json_out["return_val"] = hook(**hook_input["kwargs"])
File "/root/.local/share/pypoetry/venv/lib64/python3.9/site-packages/pyproject_hooks/_in_process/_in_process.py", line 134, in get_requires_for_build_wheel
return hook(config_settings)
File "/tmp/tmpam4wz8v1/.venv/lib/python3.9/site-packages/setuptools/build_meta.py", line 327, in get_requires_for_build_wheel
return self._get_build_requires(config_settings, requirements=[])
File "/tmp/tmpam4wz8v1/.venv/lib/python3.9/site-packages/setuptools/build_meta.py", line 297, in _get_build_requires
self.run_setup()
File "/tmp/tmpam4wz8v1/.venv/lib/python3.9/site-packages/setuptools/build_meta.py", line 497, in run_setup
super().run_setup(setup_script=setup_script)
File "/tmp/tmpam4wz8v1/.venv/lib/python3.9/site-packages/setuptools/build_meta.py", line 313, in run_setup
exec(code, locals())
File "<string>", line 9, in <module>
ImportError: cannot import name 'convert_path' from 'setuptools' (/tmp/tmpam4wz8v1/.venv/lib/python3.9/site-packages/setuptools/__init__.py)
at ~/.local/share/pypoetry/venv/lib64/python3.9/site-packages/poetry/installation/chef.py:164 in _prepare
160│
161│ error = ChefBuildError("\n\n".join(message_parts))
162│
163│ if error is not None:
→ 164│ raise error from None
165│
166│ return path
167│
168│ def _prepare_sdist(self, archive: Path, destination: Path | None = None) -> Path:
Note: This error originates from the build backend, and is likely not a problem with poetry but with django-allauth (0.45.0) not supporting PEP 517 builds. You can verify this by running 'pip wheel --no-cache-dir --use-pep517 "django-allauth (==0.45.0)"'.
```
~~I was able to install django-allauth manually using pip, but that doesn't prevent poetry from trying to install it. I couldn't find a requirements.py file to remove it as a dependency.~~
|
closed
|
2024-07-16T15:56:46Z
|
2024-07-20T21:49:57Z
|
https://github.com/milesmcc/shynet/issues/329
|
[] |
CarlSinclair
| 3
|
pydata/xarray
|
pandas
| 9,608
|
xarray can open a nc file with open_dataset, but fails to load this nc file with load
|
### What is your issue?
Recently, I have downloaded chla data from copernicus marine service, and tried to regrid it with xarray. The sad thing is that the data always goes wrong in the load phase. I have checked that variables in test dataset could be plotted normally. I do know what happen to this. Any advice is appreciated.
The test code:
```
import xarray as xr
ds = xr.open_dataset("chla201601.nc")
ds.load()
```
Test dataset:
[chla201601.zip](https://github.com/user-attachments/files/17341303/chla201601.zip)
Error information:
<details><summary>Details</summary>
<p>
```python-traceback
---------------------------------------------------------------------------
RuntimeError Traceback (most recent call last)
Cell In[3], line 1
----> 1 ds.load()
File [/usr/miniforge3/envs/xesmf_env/lib/python3.12/site-packages/xarray/core/dataset.py:880](http://localhost:8888/usr/miniforge3/envs/xesmf_env/lib/python3.12/site-packages/xarray/core/dataset.py#line=879), in Dataset.load(self, **kwargs)
878 for k, v in self.variables.items():
879 if k not in lazy_data:
--> 880 v.load()
882 return self
File [/usr/miniforge3/envs/xesmf_env/lib/python3.12/site-packages/xarray/core/variable.py:981](http://localhost:8888/usr/miniforge3/envs/xesmf_env/lib/python3.12/site-packages/xarray/core/variable.py#line=980), in Variable.load(self, **kwargs)
964 def load(self, **kwargs):
965 """Manually trigger loading of this variable's data from disk or a
966 remote source into memory and return this variable.
967
(...)
979 dask.array.compute
980 """
--> 981 self._data = to_duck_array(self._data, **kwargs)
982 return self
File [/usr/miniforge3/envs/xesmf_env/lib/python3.12/site-packages/xarray/namedarray/pycompat.py:134](http://localhost:8888/usr/miniforge3/envs/xesmf_env/lib/python3.12/site-packages/xarray/namedarray/pycompat.py#line=133), in to_duck_array(data, **kwargs)
131 return loaded_data
133 if isinstance(data, ExplicitlyIndexed):
--> 134 return data.get_duck_array() # type: ignore[no-untyped-call, no-any-return]
135 elif is_duck_array(data):
136 return data
File [/usr/miniforge3/envs/xesmf_env/lib/python3.12/site-packages/xarray/core/indexing.py:837](http://localhost:8888/usr/miniforge3/envs/xesmf_env/lib/python3.12/site-packages/xarray/core/indexing.py#line=836), in MemoryCachedArray.get_duck_array(self)
836 def get_duck_array(self):
--> 837 self._ensure_cached()
838 return self.array.get_duck_array()
File [/usr/miniforge3/envs/xesmf_env/lib/python3.12/site-packages/xarray/core/indexing.py:831](http://localhost:8888/usr/miniforge3/envs/xesmf_env/lib/python3.12/site-packages/xarray/core/indexing.py#line=830), in MemoryCachedArray._ensure_cached(self)
830 def _ensure_cached(self):
--> 831 self.array = as_indexable(self.array.get_duck_array())
File [/usr/miniforge3/envs/xesmf_env/lib/python3.12/site-packages/xarray/core/indexing.py:788](http://localhost:8888/usr/miniforge3/envs/xesmf_env/lib/python3.12/site-packages/xarray/core/indexing.py#line=787), in CopyOnWriteArray.get_duck_array(self)
787 def get_duck_array(self):
--> 788 return self.array.get_duck_array()
File [/usr/miniforge3/envs/xesmf_env/lib/python3.12/site-packages/xarray/core/indexing.py:651](http://localhost:8888/usr/miniforge3/envs/xesmf_env/lib/python3.12/site-packages/xarray/core/indexing.py#line=650), in LazilyIndexedArray.get_duck_array(self)
647 array = apply_indexer(self.array, self.key)
648 else:
649 # If the array is not an ExplicitlyIndexedNDArrayMixin,
650 # it may wrap a BackendArray so use its __getitem__
--> 651 array = self.array[self.key]
653 # self.array[self.key] is now a numpy array when
654 # self.array is a BackendArray subclass
655 # and self.key is BasicIndexer((slice(None, None, None),))
656 # so we need the explicit check for ExplicitlyIndexed
657 if isinstance(array, ExplicitlyIndexed):
File [/usr/miniforge3/envs/xesmf_env/lib/python3.12/site-packages/xarray/backends/netCDF4_.py:100](http://localhost:8888/usr/miniforge3/envs/xesmf_env/lib/python3.12/site-packages/xarray/backends/netCDF4_.py#line=99), in NetCDF4ArrayWrapper.__getitem__(self, key)
99 def __getitem__(self, key):
--> 100 return indexing.explicit_indexing_adapter(
101 key, self.shape, indexing.IndexingSupport.OUTER, self._getitem
102 )
File [/usr/miniforge3/envs/xesmf_env/lib/python3.12/site-packages/xarray/core/indexing.py:1015](http://localhost:8888/usr/miniforge3/envs/xesmf_env/lib/python3.12/site-packages/xarray/core/indexing.py#line=1014), in explicit_indexing_adapter(key, shape, indexing_support, raw_indexing_method)
993 """Support explicit indexing by delegating to a raw indexing method.
994
995 Outer and[/or](http://localhost:8888/or) vectorized indexers are supported by indexing a second time
(...)
1012 Indexing result, in the form of a duck numpy-array.
1013 """
1014 raw_key, numpy_indices = decompose_indexer(key, shape, indexing_support)
-> 1015 result = raw_indexing_method(raw_key.tuple)
1016 if numpy_indices.tuple:
1017 # index the loaded np.ndarray
1018 indexable = NumpyIndexingAdapter(result)
File [/usr/miniforge3/envs/xesmf_env/lib/python3.12/site-packages/xarray/backends/netCDF4_.py:113](http://localhost:8888/usr/miniforge3/envs/xesmf_env/lib/python3.12/site-packages/xarray/backends/netCDF4_.py#line=112), in NetCDF4ArrayWrapper._getitem(self, key)
111 with self.datastore.lock:
112 original_array = self.get_array(needs_lock=False)
--> 113 array = getitem(original_array, key)
114 except IndexError:
115 # Catch IndexError in netCDF4 and return a more informative
116 # error message. This is most often called when an unsorted
117 # indexer is used before the data is loaded from disk.
118 msg = (
119 "The indexing operation you are attempting to perform "
120 "is not valid on netCDF4.Variable object. Try loading "
121 "your data into memory first by calling .load()."
122 )
File src[/netCDF4/_netCDF4.pyx:4981](http://localhost:8888/netCDF4/_netCDF4.pyx#line=4980), in netCDF4._netCDF4.Variable.__getitem__()
File src[/netCDF4/_netCDF4.pyx:5953](http://localhost:8888/netCDF4/_netCDF4.pyx#line=5952), in netCDF4._netCDF4.Variable._get()
File src[/netCDF4/_netCDF4.pyx:2113](http://localhost:8888/netCDF4/_netCDF4.pyx#line=2112), in netCDF4._netCDF4._ensure_nc_success()
RuntimeError: NetCDF: HDF error
```
</p>
</details>
Main package information:
> xarray 2024.9.0
> numpy 2.0.2
> netCDF 4 1.7.1
> h5netcdf 1.4.0
> python 3.12.7
The ram information:
total used free shared buff/cache available
Mem: 8.3Gi 2.1Gi 5.3Gi 45Mi 1.2Gi 6.2Gi
Swap: 3.9Gi 0B 3.9Gi
|
open
|
2024-10-11T10:29:44Z
|
2024-10-25T01:33:57Z
|
https://github.com/pydata/xarray/issues/9608
|
[
"needs info"
] |
onion5376
| 3
|
TencentARC/GFPGAN
|
pytorch
| 564
|
Problem about finetuning GFPGAN v1.4
|
Hello Xintao,
We found that the direct inferencing based on GFPGAN v1.4 performs pretty well on our own datasets, whilst GFPGAN v1 inferencing is not high-quality.
However, when we tried to finetune this model, we found that only the related files of training GFPGAN v1 was released.
Could you please share the .pth files, including discriminator, with us? So l could finetune the GFPGAN v1.4 with our custom data.
Thank you very much!
Best
Junheng
|
open
|
2024-08-03T09:29:59Z
|
2024-08-23T10:33:21Z
|
https://github.com/TencentARC/GFPGAN/issues/564
|
[] |
leonsylarfang
| 4
|
mars-project/mars
|
numpy
| 3,211
|
[BUG] Ray executor auto merge chunk may raise KeyError
|
<!--
Thank you for your contribution!
Please review https://github.com/mars-project/mars/blob/master/CONTRIBUTING.rst before opening an issue.
-->
**Describe the bug**
A clear and concise description of what the bug is.
```python
mars/deploy/oscar/tests/test_ray_dag.py:189 (test_merge_groupby[before-None])
ray_start_regular_shared2 = RayContext(dashboard_url='', python_version='3.8.13', ray_version='1.13.0', ray_commit='e4ce38d001dbbe09cd21c497fedd03...127.0.0.1:64894', 'address': '127.0.0.1:64894', 'node_id': '987c20539d0bb8031ea7d8ddfc5783c01d5b79d143191bdb072ba21b'})
create_cluster = (<mars.deploy.oscar.local.LocalClient object at 0x31b18edc0>, {})
method = None, auto_merge = 'before'
@require_ray
@pytest.mark.parametrize("method", ["broadcast", None])
@pytest.mark.parametrize("auto_merge", ["before", "after"])
def test_merge_groupby(ray_start_regular_shared2, create_cluster, method, auto_merge):
rs = np.random.RandomState(0)
raw1 = pd.DataFrame({"a": rs.randint(3, size=100), "b": rs.rand(100)})
raw2 = pd.DataFrame({"a": rs.randint(3, size=10), "c": rs.rand(10)})
df1 = md.DataFrame(raw1, chunk_size=10).execute()
df2 = md.DataFrame(raw2, chunk_size=10).execute()
# do not trigger auto merge
df3 = df1.merge(
df2, on="a", auto_merge_threshold=8, method=method, auto_merge=auto_merge
)
df4 = df3.groupby("a").sum()
> result = df4.execute().fetch()
mars/deploy/oscar/tests/test_ray_dag.py:205:
_ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _
mars/core/entity/tileables.py:462: in execute
result = self.data.execute(session=session, **kw)
mars/core/entity/executable.py:144: in execute
return execute(self, session=session, **kw)
mars/deploy/oscar/session.py:1890: in execute
return session.execute(
mars/deploy/oscar/session.py:1684: in execute
execution_info: ExecutionInfo = fut.result(
../../.pyenv/versions/3.8.13/lib/python3.8/concurrent/futures/_base.py:444: in result
return self.__get_result()
../../.pyenv/versions/3.8.13/lib/python3.8/concurrent/futures/_base.py:389: in __get_result
raise self._exception
mars/deploy/oscar/session.py:1870: in _execute
await execution_info
mars/deploy/oscar/session.py:105: in wait
return await self._aio_task
mars/deploy/oscar/session.py:953: in _run_in_background
raise task_result.error.with_traceback(task_result.traceback)
mars/services/task/supervisor/processor.py:368: in run
async for stage_args in self._iter_stage_chunk_graph():
mars/services/task/supervisor/processor.py:158: in _iter_stage_chunk_graph
chunk_graph = await self._get_next_chunk_graph(chunk_graph_iter)
mars/services/task/supervisor/processor.py:149: in _get_next_chunk_graph
chunk_graph = await fut
mars/lib/aio/_threads.py:36: in to_thread
return await loop.run_in_executor(None, func_call)
../../.pyenv/versions/3.8.13/lib/python3.8/concurrent/futures/thread.py:57: in run
result = self.fn(*self.args, **self.kwargs)
mars/services/task/supervisor/processor.py:144: in next_chunk_graph
return next(chunk_graph_iter)
mars/services/task/supervisor/preprocessor.py:194: in tile
for chunk_graph in chunk_graph_builder.build():
mars/core/graph/builder/chunk.py:440: in build
yield from self._build()
mars/core/graph/builder/chunk.py:434: in _build
graph = next(tile_iterator)
mars/services/task/supervisor/preprocessor.py:74: in _iter_without_check
to_update_tileables = self._iter()
mars/core/graph/builder/chunk.py:317: in _iter
self._tile(
mars/core/graph/builder/chunk.py:211: in _tile
need_process = next(tile_handler)
mars/core/graph/builder/chunk.py:183: in _tile_handler
tiled_tileables = yield from handler.tile(tiled_tileables)
mars/core/entity/tileables.py:79: in tile
tiled_result = yield from tile_handler(op)
mars/dataframe/merge/merge.py:729: in tile
left = auto_merge_chunks(ctx, left)
mars/dataframe/utils.py:1355: in auto_merge_chunks
metas = ctx.get_chunks_meta(
_ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _
self = <mars.services.task.execution.ray.context.RayExecutionContext object at 0x31c7432b0>
data_keys = ['ed0cb85eeb0149649a565523a17aee60_0', 'ef36ff532158e2c4219867243b37f2dd_0', 'd9f91608f2ca6d88396d91ebdd9ff435_0', 'dc92a54294b3a665971b5b15da6ddd0b_0', 'c7cbea6d90a45df0826bc2a267b72d15_0', 'f769e2009ccc91538652404889dcf893_0', ...]
fields = ['memory_size'], error = 'ignore'
@implements(Context.get_chunks_meta)
def get_chunks_meta(
self, data_keys: List[str], fields: List[str] = None, error="raise"
) -> List[Dict]:
result = []
# TODO(fyrestone): Support get_chunks_meta from meta service if needed.
for key in data_keys:
> chunk_meta = self._task_chunks_meta[key]
E KeyError: 'ed0cb85eeb0149649a565523a17aee60_0'
mars/services/task/execution/ray/context.py:141: KeyError
```
**To Reproduce**
To help us reproducing this bug, please provide information below:
1. Your Python version 3.8.13
2. The version of Mars you use Latest master
3. Versions of crucial packages, such as numpy, scipy and pandas
4. Full stack of the error.
5. Minimized code to reproduce the error.
**Expected behavior**
A clear and concise description of what you expected to happen.
**Additional context**
Add any other context about the problem here.
|
closed
|
2022-08-09T02:51:44Z
|
2022-08-19T03:30:10Z
|
https://github.com/mars-project/mars/issues/3211
|
[
"type: bug",
"mod: ray integration"
] |
fyrestone
| 0
|
vimalloc/flask-jwt-extended
|
flask
| 502
|
Jwt_required and optional == True
|
This appears to be a bug unless I'm missing the requirement/intended functionality: If optional is True, I'm expecting jwt-extended not to raise any errors - however, I'm still getting a 401 - token missing error. I dug in a bit deeper in the code, and the issue seems to be that the verify_jwt_in_request method's error handler only catches NoAuthorizationError. However, _decode_jwt_from_config reraises ExpiredSignatureError....
stack:
jwt_required()
verify_jwt_in_request
_decode_jwt_from_request
decode_token
jwt_manager._decode_jwt_from_config <==
The only way to fix this is by adding the exception from the jwt manager to the verify_jwt_in_request method like so:
```
except (NoAuthorizationError, ExpiredSignatureError):
if not optional:
raise
g._jwt_extended_jwt = {}
g._jwt_extended_jwt_header = {}
g._jwt_extended_jwt_user = {"loaded_user": None}
g._jwt_extended_jwt_location = None
return None
```
Let me know if this makes sense
|
closed
|
2022-11-15T19:08:50Z
|
2022-12-22T22:52:30Z
|
https://github.com/vimalloc/flask-jwt-extended/issues/502
|
[] |
hooverdirt
| 1
|
sktime/sktime
|
scikit-learn
| 7,652
|
[BUG] AttributeError in SubLOF with novelty=False when calling fit_transform or fit_predict
|
When using SubLOF from the sktime library with novelty=False and calling fit_transform or fit_predict, an AttributeError is raised. The error indicates that the predict method is not available when novelty=False, which contradicts the documentation's recommendation to use fit_predict for outlier detection on the training data.
**To Reproduce**
```python
import pandas as pd
from sktime.annotation.lof import SubLOF
model = SubLOF(3, window_size=5, novelty=False)
x = pd.DataFrame([0, 0.5, 100, 0.1, 0, 0, 0, 100, 0, 0, 0.3, -1, 0, 100, 0.2])
model.fit_transform(x)
```
**Expected behavior**
I expected the fit_predict method to detect outliers in the provided dataset x without raising an AttributeError. According to the documentation, fit_predict should be used for outlier detection when novelty=False.
From the documentation:
> By default, LocalOutlierFactor is only meant to be used for outlier
> detection (novelty=False). Set novelty to True if you want to use
> LocalOutlierFactor for novelty detection. In this case be aware that
> you should only use predict, decision_function and score_samples
> on new unseen data and not on the training set; and note that the
> results obtained this way may differ from the standard LOF results.
**Additional context**
When 'novelty=True' is set, a warning is raised:
UserWarning: Warning: the Y parameter in detection/annotation algorithms is deprecated and will be removed in the 0.37.0 release. Users should use the y parameter instead. The class SubLOF uses the Y parameter internally in _fit, this should be replaced with y by a maintainer. Until the 0.37.0 release, this will raise no exceptions, ensuring backwards compatibility.
warn(
This is due to passing Y=None to the default function:
```python
def fit_transform(self, X, y=None, Y=None):
```
**Versions**
<details>
System:
python: 3.10.11 (v3.10.11:7d4cc5aa85, Apr 4 2023, 19:05:19) [Clang 13.0.0 (clang-1300.0.29.30)]
executable: /usr/local/bin/python3
machine: macOS-15.2-arm64-arm-64bit
Python dependencies:
pip: 24.3.1
sktime: 0.35.0
sklearn: 1.5.2
skbase: 0.12.0
numpy: 1.26.4
scipy: 1.14.1
pandas: 2.2.3
matplotlib: 3.9.3
joblib: 1.4.2
numba: 0.60.0
statsmodels: 0.14.4
pmdarima: None
statsforecast: 2.0.0
tsfresh: None
tslearn: None
torch: 2.5.1
tensorflow: 2.16.2
</details>
|
open
|
2025-01-17T07:44:33Z
|
2025-03-15T23:17:41Z
|
https://github.com/sktime/sktime/issues/7652
|
[
"bug",
"module:detection"
] |
axisrow
| 4
|
ryfeus/lambda-packs
|
numpy
| 21
|
Lamda Packs for Tensorflow 1.6 based on Python 3
|
Hi, I'm trying to create a lamda pack for tensorflow 1.6 based on Python 3.
However I am not able to compress the pack to be less than 50 MB.
Can you please share your taught's on how to do that.
|
closed
|
2018-05-23T07:28:04Z
|
2018-12-13T18:13:50Z
|
https://github.com/ryfeus/lambda-packs/issues/21
|
[] |
SumanthReddyKaliki
| 1
|
aminalaee/sqladmin
|
fastapi
| 172
|
Be able to hook into the fastAPI subapp
|
### Checklist
- [X] There are no similar issues or pull requests for this yet.
### Is your feature related to a problem? Please describe.
Hello, great work ! :)
I would like to be able to add my own url paths below the root endpoint of the admin app.
For example I would like to be able to do a POST on the homepage (url "/")
Something that would look like this
```python
@router.post("")
async def post_homepage(request: Request):
logger.warning("post !!")
return templates.TemplateResponse("index.html", {"request": request})
```
From what I understand it is not possible to access or modify the sub app
Neither is it possible to mount another sub application on the same endpoint (because routes would conflict)
It would be great if we could use the default routes but also add some and override existing routes
### Describe the solution you would like.
```python
from backoffice import homepage
admin = Admin(app, create_engine(Settings().db_uri), base_url="/backoffice")
admin.include_router(homepage.router)
```
routes could be added before the existing routes to have priority
### Describe alternatives you considered
_No response_
### Additional context
_No response_
|
open
|
2022-06-08T16:18:35Z
|
2022-06-24T12:18:34Z
|
https://github.com/aminalaee/sqladmin/issues/172
|
[
"waiting-for-feedback"
] |
lebrunthibault
| 5
|
feder-cr/Jobs_Applier_AI_Agent_AIHawk
|
automation
| 347
|
Phone number and country did not change
|
I have changed the phone number and the country in plain_text_resume.yaml but when i see the bot applying to jobs , It write italy in the country and different phone number.
I apply with my own pdf using python main.py --resume /path/to/your/resume.pdf

|
closed
|
2024-09-10T16:32:50Z
|
2024-09-13T11:53:22Z
|
https://github.com/feder-cr/Jobs_Applier_AI_Agent_AIHawk/issues/347
|
[] |
db2024git
| 2
|
aiortc/aiortc
|
asyncio
| 200
|
example server not working on remote machine
|
The example server code works perfectly locally, even within a docker.
However, if I run the server code on a remote machine, and open the webpage from the local machine browser, ICE gathering state is always new.
Did I miss anything? Or it indeed takes very long to connect?
Much appreciate it.
|
closed
|
2019-08-04T04:26:14Z
|
2019-08-17T06:09:33Z
|
https://github.com/aiortc/aiortc/issues/200
|
[] |
litanlitudan
| 2
|
ultrafunkamsterdam/undetected-chromedriver
|
automation
| 840
|
Give Old Docker image Dockerfile
|
Recently you updated your docker image. Could you give me the old dockerfile as the newer one takes a lot of time to build as it installs chrome.
|
closed
|
2022-10-15T16:42:07Z
|
2022-10-16T07:18:32Z
|
https://github.com/ultrafunkamsterdam/undetected-chromedriver/issues/840
|
[] |
Chetan11-dev
| 2
|
adamerose/PandasGUI
|
pandas
| 168
|
Change the qt calls to use qtpy to deal with different Qt bindings
|
Today the best approach to ensure some consistency and flexibility across different Qt python bindings (PyQt5, PySide2, PyQt6, PySide6, ...) is the use of [QtPy: Abstraction layer for PyQt5/PyQt4/PySide2/PySide](https://github.com/spyder-ide/qtpy).
I propose a set of minor changes to ensure all the calls to Qt bindings are performed via from qtpy import.
By default the module to be called will be PyQt5 but it can be changed by setting the environment variable QT_API.
If the proposal is accepted, I will submit a git commit against the latest version.
|
open
|
2021-08-29T08:44:43Z
|
2021-08-29T21:19:40Z
|
https://github.com/adamerose/PandasGUI/issues/168
|
[] |
EmanueleCannizzaro
| 1
|
mljar/mercury
|
data-visualization
| 417
|
Option to hide the RunMercury switch at the top black bar
|
Hello:
I have been testing this product and it's wonderful... Only issues I had are with:
- Share button: in some cases, I would prefer not to have our users sharing the links for security reasons.
- The RunMercury switch at the top blank bar, might not be good in those cases where you don't want the users to check all available notebooks, so it would be good to have an option to hide/show this.
Are there these options considered in this product's roadmap?
Regards;
Greg
|
closed
|
2024-02-04T23:17:10Z
|
2024-02-15T13:37:14Z
|
https://github.com/mljar/mercury/issues/417
|
[] |
gregcode123
| 3
|
MorvanZhou/tutorials
|
tensorflow
| 78
|
建议
|
建议课程中多讲一点理论,比如数学基础等。感谢提供教程
|
closed
|
2019-03-08T03:01:44Z
|
2019-03-08T03:03:35Z
|
https://github.com/MorvanZhou/tutorials/issues/78
|
[] |
ustcerlik
| 0
|
jschneier/django-storages
|
django
| 869
|
""Unicode-objects must be encoded before hashing" when i try upload a .m3u8 file using storages|s3 instead local storage.
|
**Describe the bug**
**COMPLETE CODE AND SETTINGS IN THE END OF THE FILE**
**the upload works when i add the .m3u8 file directly in the aws s3 site**
I was able to send the video normally to my local machine, but when I changed the storage settings, I just started getting this error.
The error points to the line where the code is
```
instance.file.save(file_name_m3u8, file_m3u8)
```
And then immediately points to
```
.../python3.8/site-packages/storages/backends/s3boto3.py
...
obj.upload_fileobj(content, ExtraArgs=params)
...
```
My file object `file_m3u8` is:
```
file_object
<_io.TextIOWrapper name='/tmp/media/lectures/first_video_2/2020-04-04_16-11-20.m3u8' mode='r' encoding='UTF-8'>
```
Example:
```
from django.db.models.signals import post_save
from django.dispatch import receiver
from django.core.files import File
from .models import Lecture
@receiver(post_save, sender=Lecture)
def handle_video_upload (sender, instance, created, ** kwargs):
with open (
"/tmp/media/lectures/first_video_2/2020-04-04_16-11-20.m3u8", "r") as file_object:
file_m3u8 = File (
name = "media/lectures/first_video_2/2020-04-04_16-11-20.m3u8",
file = file_object)
instance.file.save ("2020-04-04_16-11-20.m3u8", file_m3u8)
```
I'm using django-storages and settings this in my settings.py file:
```
STATICFILES_STORAGE = "django.contrib.staticfiles.storage.ManifestStaticFilesStorage"
DEFAULT_FILE_STORAGE = "storages.backends.s3boto3.S3Boto3Storage"
AWS_ACCESS_KEY_ID = config("AWS_ACCESS_KEY_ID")
AWS_SECRET_ACCESS_KEY = config("AWS_SECRET_ACCESS_KEY")
AWS_STORAGE_BUCKET_NAME = config("AWS_STORAGE_BUCKET_NAME")
CLOUDFRONT_ID = config("CLOUDFRONT_ID")
CLOUDFRONT_DOMAIN = f"{CLOUDFRONT_ID}.cloudfront.net"
AWS_S3_CUSTOM_DOMAIN = f"{CLOUDFRONT_ID}.cloudfront.net"
```
**Expected behavior**
i expect that the file will be uploaded normally
**Debug logs**
terminal output:
```
[05/Apr/2020 17:42:30] "GET /admin/jsi18n/ HTTP/1.1" 200 7275
/home/marcos/.local/share/virtualenvs/geeknoon_server-HmoJlbqP/lib/python3.8/site-packages/storages/backends/s3boto3.py:340: UserWarning: The default behavior of S3Boto3Storage is insecure and will change in django-storages 1.10. By default files and new buckets are saved with an ACL of 'public-read' (globally publicly readable). Version 1.10 will default to using the bucket's ACL. To opt into the new behavior set AWS_DEFAULT_ACL = None, otherwise to silence this warning explicitly set AWS_DEFAULT_ACL.
warnings.warn(
[05/Apr/2020 17:42:32] "GET /admin/lectures/lecture/5/change/ HTTP/1.1" 200 6712
[05/Apr/2020 17:42:32] "GET /admin/jsi18n/ HTTP/1.1" 200 7275
/tmp/media/lectures/first_video_2/2020-04-04_16-11-20.m3u8
2020-04-04_16-11-20.m3u8
Internal Server Error: /admin/lectures/lecture/5/change/
Traceback (most recent call last):
File "/home/marcos/.local/share/virtualenvs/geeknoon_server-HmoJlbqP/lib/python3.8/site-packages/django/core/handlers/exception.py", line 34, in inner
response = get_response(request)
File "/home/marcos/.local/share/virtualenvs/geeknoon_server-HmoJlbqP/lib/python3.8/site-packages/django/core/handlers/base.py", line 115, in _get_response
response = self.process_exception_by_middleware(e, request)
File "/home/marcos/.local/share/virtualenvs/geeknoon_server-HmoJlbqP/lib/python3.8/site-packages/django/core/handlers/base.py", line 113, in _get_response
response = wrapped_callback(request, *callback_args, **callback_kwargs)
File "/home/marcos/.local/share/virtualenvs/geeknoon_server-HmoJlbqP/lib/python3.8/site-packages/django/contrib/admin/options.py", line 607, in wrapper
return self.admin_site.admin_view(view)(*args, **kwargs)
File "/home/marcos/.local/share/virtualenvs/geeknoon_server-HmoJlbqP/lib/python3.8/site-packages/django/utils/decorators.py", line 130, in _wrapped_view
response = view_func(request, *args, **kwargs)
File "/home/marcos/.local/share/virtualenvs/geeknoon_server-HmoJlbqP/lib/python3.8/site-packages/django/views/decorators/cache.py", line 44, in _wrapped_view_func
response = view_func(request, *args, **kwargs)
File "/home/marcos/.local/share/virtualenvs/geeknoon_server-HmoJlbqP/lib/python3.8/site-packages/django/contrib/admin/sites.py", line 231, in inner
return view(request, *args, **kwargs)
File "/home/marcos/.local/share/virtualenvs/geeknoon_server-HmoJlbqP/lib/python3.8/site-packages/django/contrib/admin/options.py", line 1641, in change_view
return self.changeform_view(request, object_id, form_url, extra_context)
File "/home/marcos/.local/share/virtualenvs/geeknoon_server-HmoJlbqP/lib/python3.8/site-packages/django/utils/decorators.py", line 43, in _wrapper
return bound_method(*args, **kwargs)
File "/home/marcos/.local/share/virtualenvs/geeknoon_server-HmoJlbqP/lib/python3.8/site-packages/django/utils/decorators.py", line 130, in _wrapped_view
response = view_func(request, *args, **kwargs)
File "/home/marcos/.local/share/virtualenvs/geeknoon_server-HmoJlbqP/lib/python3.8/site-packages/django/contrib/admin/options.py", line 1522, in changeform_view
return self._changeform_view(request, object_id, form_url, extra_context)
File "/home/marcos/.local/share/virtualenvs/geeknoon_server-HmoJlbqP/lib/python3.8/site-packages/django/contrib/admin/options.py", line 1565, in _changeform_view
self.save_model(request, new_object, form, not add)
File "/home/marcos/.local/share/virtualenvs/geeknoon_server-HmoJlbqP/lib/python3.8/site-packages/django/contrib/admin/options.py", line 1081, in save_model
obj.save()
File "/home/marcos/.local/share/virtualenvs/geeknoon_server-HmoJlbqP/lib/python3.8/site-packages/django/db/models/base.py", line 745, in save
self.save_base(using=using, force_insert=force_insert,
File "/home/marcos/.local/share/virtualenvs/geeknoon_server-HmoJlbqP/lib/python3.8/site-packages/django/db/models/base.py", line 793, in save_base
post_save.send(
File "/home/marcos/.local/share/virtualenvs/geeknoon_server-HmoJlbqP/lib/python3.8/site-packages/django/dispatch/dispatcher.py", line 173, in send
return [
File "/home/marcos/.local/share/virtualenvs/geeknoon_server-HmoJlbqP/lib/python3.8/site-packages/django/dispatch/dispatcher.py", line 174, in <listcomp>
(receiver, receiver(signal=self, sender=sender, **named))
File "/home/marcos/geeknoon/geeknoon_server/lectures/signals.py", line 90, in handle_video_upload
instance.file.save(file_name_m3u8, file_m3u8)
File "/home/marcos/.local/share/virtualenvs/geeknoon_server-HmoJlbqP/lib/python3.8/site-packages/django/db/models/fields/files.py", line 87, in save
self.name = self.storage.save(name, content, max_length=self.field.max_length)
File "/home/marcos/.local/share/virtualenvs/geeknoon_server-HmoJlbqP/lib/python3.8/site-packages/django/core/files/storage.py", line 52, in save
return self._save(name, content)
File "/home/marcos/.local/share/virtualenvs/geeknoon_server-HmoJlbqP/lib/python3.8/site-packages/storages/backends/s3boto3.py", line 547, in _save
obj.upload_fileobj(content, ExtraArgs=params)
File "/home/marcos/.local/share/virtualenvs/geeknoon_server-HmoJlbqP/lib/python3.8/site-packages/boto3/s3/inject.py", line 619, in object_upload_fileobj
return self.meta.client.upload_fileobj(
File "/home/marcos/.local/share/virtualenvs/geeknoon_server-HmoJlbqP/lib/python3.8/site-packages/boto3/s3/inject.py", line 539, in upload_fileobj
return future.result()
File "/home/marcos/.local/share/virtualenvs/geeknoon_server-HmoJlbqP/lib/python3.8/site-packages/s3transfer/futures.py", line 106, in result
return self._coordinator.result()
File "/home/marcos/.local/share/virtualenvs/geeknoon_server-HmoJlbqP/lib/python3.8/site-packages/s3transfer/futures.py", line 265, in result
raise self._exception
File "/home/marcos/.local/share/virtualenvs/geeknoon_server-HmoJlbqP/lib/python3.8/site-packages/s3transfer/tasks.py", line 126, in __call__
return self._execute_main(kwargs)
File "/home/marcos/.local/share/virtualenvs/geeknoon_server-HmoJlbqP/lib/python3.8/site-packages/s3transfer/tasks.py", line 150, in _execute_main
return_value = self._main(**kwargs)
File "/home/marcos/.local/share/virtualenvs/geeknoon_server-HmoJlbqP/lib/python3.8/site-packages/s3transfer/upload.py", line 692, in _main
client.put_object(Bucket=bucket, Key=key, Body=body, **extra_args)
File "/home/marcos/.local/share/virtualenvs/geeknoon_server-HmoJlbqP/lib/python3.8/site-packages/botocore/client.py", line 316, in _api_call
return self._make_api_call(operation_name, kwargs)
File "/home/marcos/.local/share/virtualenvs/geeknoon_server-HmoJlbqP/lib/python3.8/site-packages/botocore/client.py", line 602, in _make_api_call
handler, event_response = self.meta.events.emit_until_response(
File "/home/marcos/.local/share/virtualenvs/geeknoon_server-HmoJlbqP/lib/python3.8/site-packages/botocore/hooks.py", line 360, in emit_until_response
return self._emitter.emit_until_response(aliased_event_name, **kwargs)
File "/home/marcos/.local/share/virtualenvs/geeknoon_server-HmoJlbqP/lib/python3.8/site-packages/botocore/hooks.py", line 243, in emit_until_response
responses = self._emit(event_name, kwargs, stop_on_response=True)
File "/home/marcos/.local/share/virtualenvs/geeknoon_server-HmoJlbqP/lib/python3.8/site-packages/botocore/hooks.py", line 211, in _emit
response = handler(**kwargs)
File "/home/marcos/.local/share/virtualenvs/geeknoon_server-HmoJlbqP/lib/python3.8/site-packages/botocore/handlers.py", line 216, in conditionally_calculate_md5
calculate_md5(params, **kwargs)
File "/home/marcos/.local/share/virtualenvs/geeknoon_server-HmoJlbqP/lib/python3.8/site-packages/botocore/handlers.py", line 194, in calculate_md5
binary_md5 = _calculate_md5_from_file(body)
File "/home/marcos/.local/share/virtualenvs/geeknoon_server-HmoJlbqP/lib/python3.8/site-packages/botocore/handlers.py", line 208, in _calculate_md5_from_file
md5.update(chunk)
TypeError: Unicode-objects must be encoded before hashing
[05/Apr/2020 17:42:39] "POST /admin/lectures/lecture/5/change/ HTTP/1.1" 500 249595
```
## Complete code in signals.py:
```
import subprocess
import os
from django.db.models.signals import post_save
from django.dispatch import receiver
from django.core.files import File
from pathlib import Path
from .models import Lecture
@receiver(post_save, sender=Lecture)
def handle_video_upload(sender, instance, created, **kwargs):
file_relative_path = Path(instance.file.name)
file_suffix = file_relative_path.suffix
if not file_suffix == '.m3u8' and instance.file_type == "V":
file_relative_dir = os.path.dirname(instance.file.name)
file_relative_path_m3u8 = file_relative_path.with_suffix(".m3u8")
file_name_m3u8 = file_relative_path_m3u8.name
file_tmp_local_dir = f"/tmp/{file_relative_dir}"
file_tmp_local_output = f"{file_tmp_local_dir}/{file_name_m3u8}"
file_cloudfront_url = instance.file.url
subprocess.run(['mkdir', '-p', file_tmp_local_dir])
subprocess.run([
"ffmpeg",
"-i",
file_cloudfront_url,
"-f",
"hls",
file_tmp_local_output,
'-loglevel',
'quiet'
])
<comment: update the file with the new .m3u8 file>
with open(file_tmp_local_output, "r") as file_object:
print(file_tmp_local_output)
print(file_name_m3u8)
file_m3u8 = File(name=file_relative_path_m3u8, file=file_object)
instance.file.save(file_name_m3u8, file_m3u8)
subprocess.run(["rm", "-r", file_tmp_local_dir])
boto3.set_stream_logger('')
```
All relevant settings.py vars:
```
INSTALLED_APPS = [
"storages",
]
STATICFILES_FINDERS = [
"django.contrib.staticfiles.finders.FileSystemFinder",
"django.contrib.staticfiles.finders.AppDirectoriesFinder",
]
STATIC_ROOT = BASE_DIR.joinpath("local", "static")
MEDIA_ROOT = BASE_DIR.joinpath("local", "media")
STATICFILES_STORAGE = "django.contrib.staticfiles.storage.ManifestStaticFilesStorage"
DEFAULT_FILE_STORAGE = "storages.backends.s3boto3.S3Boto3Storage"
AWS_ACCESS_KEY_ID = config("AWS_ACCESS_KEY_ID")
AWS_SECRET_ACCESS_KEY = config("AWS_SECRET_ACCESS_KEY")
AWS_STORAGE_BUCKET_NAME = config("AWS_STORAGE_BUCKET_NAME")
CLOUDFRONT_ID = config("CLOUDFRONT_ID")
CLOUDFRONT_DOMAIN = f"{CLOUDFRONT_ID}.cloudfront.net"
AWS_S3_CUSTOM_DOMAIN = f"{CLOUDFRONT_ID}.cloudfront.net"
```
What i trying?
Convert .mkv|.mp4 videos etc after upload, and set the new .m3u8 file in the file field in my model.
I check if this process is needed by checking the file_type and checking if the file was converted by checking if the suffix is the .m3u8.
My model relevant code:
```
from django.db import models
from base_models import CommomInfo # with updated, created and uuid fields
def lecture_file_path(instance, filename):
return f"media/lectures/{instance.slug}/{filename}"
class Lecture(CommomInfo):
FILE_TYPE_CHOICES = (("V", "Video"), ("P", "PDF"))
file = models.FileField(upload_to=lecture_file_path)
file_type = models.CharField(
max_length=1,
choices=FILE_TYPE_CHOICES,
default="V")
```
|
closed
|
2020-04-05T17:52:16Z
|
2020-04-05T18:17:47Z
|
https://github.com/jschneier/django-storages/issues/869
|
[] |
marcosfromrio
| 3
|
explosion/spaCy
|
data-science
| 12,982
|
RuntimeError: Error(s) in loading state_dict for RobertaModel: Unexpected key(s) in state_dict: "embeddings.position_ids".
|
<!-- NOTE: For questions or install related issues, please open a Discussion instead. -->
## How to reproduce the behaviour
```py
import spacy
nlp = spacy.load('en_core_web_trf')
```
Full traceback:
```
---------------------------------------------------------------------------
RuntimeError Traceback (most recent call last)
Cell In[7], line 1
----> 1 nlp = spacy.load('en_core_web_trf')
File /opt/conda/lib/python3.8/site-packages/spacy/__init__.py:51, in load(name, vocab, disable, enable, exclude, config)
27 def load(
28 name: Union[str, Path],
29 *,
(...)
34 config: Union[Dict[str, Any], Config] = util.SimpleFrozenDict(),
35 ) -> Language:
36 """Load a spaCy model from an installed package or a local path.
37
38 name (str): Package name or model path.
(...)
49 RETURNS (Language): The loaded nlp object.
50 """
---> 51 return util.load_model(
52 name,
53 vocab=vocab,
54 disable=disable,
55 enable=enable,
56 exclude=exclude,
57 config=config,
58 )
File /opt/conda/lib/python3.8/site-packages/spacy/util.py:465, in load_model(name, vocab, disable, enable, exclude, config)
463 return get_lang_class(name.replace("blank:", ""))()
464 if is_package(name): # installed as package
--> 465 return load_model_from_package(name, **kwargs) # type: ignore[arg-type]
466 if Path(name).exists(): # path to model data directory
467 return load_model_from_path(Path(name), **kwargs) # type: ignore[arg-type]
File /opt/conda/lib/python3.8/site-packages/spacy/util.py:501, in load_model_from_package(name, vocab, disable, enable, exclude, config)
484 """Load a model from an installed package.
485
486 name (str): The package name.
(...)
498 RETURNS (Language): The loaded nlp object.
499 """
500 cls = importlib.import_module(name)
--> 501 return cls.load(vocab=vocab, disable=disable, enable=enable, exclude=exclude, config=config)
File /opt/conda/lib/python3.8/site-packages/en_core_web_trf/__init__.py:10, in load(**overrides)
9 def load(**overrides):
---> 10 return load_model_from_init_py(__file__, **overrides)
File /opt/conda/lib/python3.8/site-packages/spacy/util.py:682, in load_model_from_init_py(init_file, vocab, disable, enable, exclude, config)
680 if not model_path.exists():
681 raise IOError(Errors.E052.format(path=data_path))
--> 682 return load_model_from_path(
683 data_path,
684 vocab=vocab,
685 meta=meta,
686 disable=disable,
687 enable=enable,
688 exclude=exclude,
689 config=config,
690 )
File /opt/conda/lib/python3.8/site-packages/spacy/util.py:547, in load_model_from_path(model_path, meta, vocab, disable, enable, exclude, config)
538 config = load_config(config_path, overrides=overrides)
539 nlp = load_model_from_config(
540 config,
541 vocab=vocab,
(...)
545 meta=meta,
546 )
--> 547 return nlp.from_disk(model_path, exclude=exclude, overrides=overrides)
File /opt/conda/lib/python3.8/site-packages/spacy/language.py:2155, in Language.from_disk(self, path, exclude, overrides)
2152 if not (path / "vocab").exists() and "vocab" not in exclude: # type: ignore[operator]
2153 # Convert to list here in case exclude is (default) tuple
2154 exclude = list(exclude) + ["vocab"]
-> 2155 util.from_disk(path, deserializers, exclude) # type: ignore[arg-type]
2156 self._path = path # type: ignore[assignment]
2157 self._link_components()
File /opt/conda/lib/python3.8/site-packages/spacy/util.py:1392, in from_disk(path, readers, exclude)
1389 for key, reader in readers.items():
1390 # Split to support file names like meta.json
1391 if key.split(".")[0] not in exclude:
-> 1392 reader(path / key)
1393 return path
File /opt/conda/lib/python3.8/site-packages/spacy/language.py:2149, in Language.from_disk.<locals>.<lambda>(p, proc)
2147 if not hasattr(proc, "from_disk"):
2148 continue
-> 2149 deserializers[name] = lambda p, proc=proc: proc.from_disk( # type: ignore[misc]
2150 p, exclude=["vocab"]
2151 )
2152 if not (path / "vocab").exists() and "vocab" not in exclude: # type: ignore[operator]
2153 # Convert to list here in case exclude is (default) tuple
2154 exclude = list(exclude) + ["vocab"]
File /opt/conda/lib/python3.8/site-packages/spacy_transformers/pipeline_component.py:416, in Transformer.from_disk(self, path, exclude)
409 self.model.attrs["set_transformer"](self.model, hf_model)
411 deserialize = {
412 "vocab": self.vocab.from_disk,
413 "cfg": lambda p: self.cfg.update(deserialize_config(p)),
414 "model": load_model,
415 }
--> 416 util.from_disk(path, deserialize, exclude) # type: ignore
417 return self
File /opt/conda/lib/python3.8/site-packages/spacy/util.py:1392, in from_disk(path, readers, exclude)
1389 for key, reader in readers.items():
1390 # Split to support file names like meta.json
1391 if key.split(".")[0] not in exclude:
-> 1392 reader(path / key)
1393 return path
File /opt/conda/lib/python3.8/site-packages/spacy_transformers/pipeline_component.py:390, in Transformer.from_disk.<locals>.load_model(p)
388 try:
389 with open(p, "rb") as mfile:
--> 390 self.model.from_bytes(mfile.read())
391 except AttributeError:
392 raise ValueError(Errors.E149) from None
File /opt/conda/lib/python3.8/site-packages/thinc/model.py:638, in Model.from_bytes(self, bytes_data)
636 msg = srsly.msgpack_loads(bytes_data)
637 msg = convert_recursive(is_xp_array, self.ops.asarray, msg)
--> 638 return self.from_dict(msg)
File /opt/conda/lib/python3.8/site-packages/thinc/model.py:676, in Model.from_dict(self, msg)
674 node.set_param(param_name, value)
675 for i, shim_bytes in enumerate(msg["shims"][i]):
--> 676 node.shims[i].from_bytes(shim_bytes)
677 return self
File /opt/conda/lib/python3.8/site-packages/spacy_transformers/layers/hf_shim.py:120, in HFShim.from_bytes(self, bytes_data)
118 filelike.seek(0)
119 device = get_torch_default_device()
--> 120 self._model.load_state_dict(torch.load(filelike, map_location=device))
121 self._model.to(device)
122 else:
File /opt/conda/lib/python3.8/site-packages/torch/nn/modules/module.py:2041, in Module.load_state_dict(self, state_dict, strict)
2036 error_msgs.insert(
2037 0, 'Missing key(s) in state_dict: {}. '.format(
2038 ', '.join('"{}"'.format(k) for k in missing_keys)))
2040 if len(error_msgs) > 0:
-> 2041 raise RuntimeError('Error(s) in loading state_dict for {}:\n\t{}'.format(
2042 self.__class__.__name__, "\n\t".join(error_msgs)))
2043 return _IncompatibleKeys(missing_keys, unexpected_keys)
RuntimeError: Error(s) in loading state_dict for RobertaModel:
Unexpected key(s) in state_dict: "embeddings.position_ids".
```
Also:
```
~$ conda list torch
# packages in environment at /opt/conda:
#
# Name Version Build Channel
efficientnet-pytorch 0.7.1 pyhd8ed1ab_1 conda-forge
pytorch 2.0.1 py3.8_cuda11.7_cudnn8.5.0_0 pytorch
pytorch-cuda 11.7 h67b0de4_0 pytorch
pytorch-lightning 2.0.1.post0 pypi_0 pypi
pytorch-mutex 1.0 cuda pytorch
rotary-embedding-torch 0.2.1 pypi_0 pypi
torchaudio 2.0.2 py38_cu117 pytorch
torchmetrics 0.11.4 pypi_0 pypi
torchtriton 2.0.0 py38 pytorch
torchvision 0.15.2 py38_cu117 pytorch
torchviz 0.0.2 pypi_0 pypi
```
## Your Environment
<!-- Include details of your environment. You can also type `python -m spacy info --markdown` and copy-paste the result here.-->
* Operating System:
* Python Version Used:
* spaCy Version Used:
* Environment Information:
```
spaCy version 3.6.1
Location /opt/conda/lib/python3.8/site-packages/spacy
Platform Linux-5.13.0-1023-aws-x86_64-with-glibc2.17
Python version 3.8.17
Pipelines en_core_web_trf (3.6.1)
```
|
closed
|
2023-09-14T14:08:37Z
|
2023-10-19T00:02:09Z
|
https://github.com/explosion/spaCy/issues/12982
|
[
"install",
"feat / transformer"
] |
dzenilee
| 6
|
zappa/Zappa
|
flask
| 775
|
[Migrated] 'exclude' setting in config excludes all occurrences also for dependencies
|
Originally from: https://github.com/Miserlou/Zappa/issues/1917 by [themmes](https://github.com/themmes)
<!--- Provide a general summary of the issue in the Title above -->
## Context
<!--- Provide a more detailed introduction to the issue itself, and why you consider it to be a bug -->
I do not understand why I am getting the following error. It seems the lambda cannot be loaded from S3 and therefore is not able to find the module to execute the function.
```
[1565266143895] [DEBUG] 2019-08-08T12:09:03.895Z ef9eb08e-f4f9-43a5-9072-c11f15be1b80 Changing event name from docs.*.cloudsearchdomain.Search.complete-section to docs.*.cloudsearch-domain.Search.complete-section
[1565266143897] The 's3' resource does not exist.
The available resources are:
-
: ResourceNotExistsError
Traceback (most recent call last):
File "/var/task/handler.py", line 602, in lambda_handler
return LambdaHandler.lambda_handler(event, context)
File "/var/task/handler.py", line 245, in lambda_handler
handler = cls()
File "/var/task/handler.py", line 102, in __init__
self.load_remote_project_archive(project_archive_path)
File "/var/task/handler.py", line 170, in load_remote_project_archive
s3 = boto_session.resource('s3')
File "/var/task/boto3/session.py", line 347, in resource
has_low_level_client)
boto3.exceptions.ResourceNotExistsError: The 's3' resource does not exist.
The available resources are:
-
[1565266201109] [DEBUG] 2019-08-08T12:10:01.109Z ef9eb08e-f4f9-43a5-9072-c11f15be1b80 Zappa Event: {'Records': [{'eventVersion': '2.1', 'eventSource': 'aws:s3', 'awsRegion': 'eu-central-1', 'eventTime': '2019-08-08T12:09:00.157Z', 'eventName': 'ObjectCreated:Put', 'userIdentity': {'principalId': 'AWS:AIDAY5TJ3FA44X4WWYBA7'}, 'requestParameters': {'sourceIPAddress': '213.127.67.154'}, 'responseElements': {'x-amz-request-id': 'B35B73FF4F75D59E', 'x-amz-id-2': 'ELkqeB94Gb17TPF12ffhVtASmEhtR7NlQO4DevDruHvA5I5DrFlln/oYPSJkDx9RO/D7MMxERKE='}, 's3': {'s3SchemaVersion': '1.0', 'configurationId': 'function-dev:sample.lambda_handler', 'bucket': {'name': 'test-function', 'ownerIdentity': {'principalId': 'A3V2HVB9FLXO6B'}, 'arn': 'arn:aws:s3:::test-function'}, 'object': {'key': 'test_5.pts.gz', 'size': 7924, 'eTag': '3b39cf26af0e2aa49b917a2771bcc068', 'sequencer': '005D4C10DC203B4EB2'}}}]}
[1565266201111] No module named 'sample': ModuleNotFoundError
Traceback (most recent call last):
File "/var/task/handler.py", line 602, in lambda_handler
return LambdaHandler.lambda_handler(event, context)
File "/var/task/handler.py", line 248, in lambda_handler
return handler.handler(event, context)
File "/var/task/handler.py", line 423, in handler
app_function = self.import_module_and_get_function(whole_function)
File "/var/task/handler.py", line 239, in import_module_and_get_function
app_module = importlib.import_module(module)
File "/var/lang/lib/python3.6/importlib/__init__.py", line 126, in import_module
return _bootstrap._gcd_import(name[level:], package, level)
File "<frozen importlib._bootstrap>", line 994, in _gcd_import
File "<frozen importlib._bootstrap>", line 971, in _find_and_load
File "<frozen importlib._bootstrap>", line 953, in _find_and_load_unlocked
ModuleNotFoundError: No module named 'sample'
```
- I verified the tar.gz is in the correct `bucket-zappa` bucket on S3
- The Zappa LambdaRole is correctly created and has access to S3
<!--- Also, please make sure that you are running Zappa _from a virtual environment_ and are using Python 2.7/3.6 -->
## Expected Behavior
<!--- Tell us what should happen -->
I expected the application to be found on S3 and the specific module.function to be ran upon the event.
## Actual Behavior
<!--- Tell us what happens instead -->
It seems the package cannot be found on S3 and when the S3 event comes in Lambda is not able to find the correct module.function to run
## Possible Fix
<!--- Not obligatory, but suggest a fix or reason for the bug -->
## Steps to Reproduce
<!--- Provide a link to a live example, or an unambiguous set of steps to -->
<!--- reproduce this bug include code to reproduce, if relevant -->
Its hard to specify how to reproduce because I have no idea what is causing this, from one day to the other my Lambda applications start raising this issue.
## Your Environment
<!--- Include as many relevant details about the environment you experienced the bug in -->
* Zappa version used: 0.48.2
* Operating System and Python version: Xenial 16.04 python3.6
* The output of `pip freeze`:
boto3==1.9.204
* Link to your project (optional):
* Your `zappa_settings.py`:
```
{
"dev": {
"project_name": "test",
"runtime": "python3.6",
"exclude": ["env","tests","data","output","notebooks", "launch.sh"],
"s3_bucket": "bucket-zappa",
"slim_handler": true,
"aws_region": "eu-central-1",
"events": [{
"function": "sample.lambda_handler",
"event_source": {
"arn": "arn:aws:s3:::test-function",
"events": ["s3:ObjectCreated:*"]
}
}],
"keep_warm": false,
"apigateway_enabled": false
}
}
```
|
closed
|
2021-02-20T12:42:15Z
|
2024-04-13T18:37:15Z
|
https://github.com/zappa/Zappa/issues/775
|
[
"no-activity",
"auto-closed"
] |
jneves
| 3
|
zappa/Zappa
|
django
| 695
|
[Migrated] Flask-like app factory support
|
Originally from: https://github.com/Miserlou/Zappa/issues/1775 by [fcicc](https://github.com/fcicc)
# Description
How to use application factories in Zappa:
* app.py
`def create_app():`
`app = Flask()`
`#define settings, db, routes, ...`
`return app`
* zappa_settings.json
`{`
`"app_function": "app.create_app"`
`}`
# GitHub Issue
#1771
|
closed
|
2021-02-20T12:33:03Z
|
2022-07-16T06:37:34Z
|
https://github.com/zappa/Zappa/issues/695
|
[
"needs-user-testing"
] |
jneves
| 1
|
hootnot/oanda-api-v20
|
rest-api
| 90
|
"Connection: Keep-Alive" question
|
This is more a question than an issue. It's about the best practices section of the developer references (http://developer.oanda.com/rest-live-v20/best-practices/).
They say that you should add a http header with the following content `Connection: Keep-Alive`. However I ran `grep -rn "Connection" .` in this repository and I found nothing. I would like to know if you consider this.
|
closed
|
2017-07-19T18:37:45Z
|
2017-07-20T07:49:16Z
|
https://github.com/hootnot/oanda-api-v20/issues/90
|
[] |
silgon
| 3
|
tqdm/tqdm
|
pandas
| 859
|
fractional total keyword can cause AssertionError in version 4.40
|
In the newest version, a fractional total can produce an AssertionError, where in previous versions it worked as expected. The documentation suggests that `total` can be a floating point value, so the error appears to be a bug. Here is a minimal reproducible example:
```python
import sys, tqdm
print(tqdm.__version__, sys.version, sys.platform)
for i in tqdm.tqdm(iterable=range(10), total=9.6):
pass
```
Here is the output I get in an older tqdm version:
```
4.32.2 3.6.9 |Anaconda, Inc.| (default, Jul 30 2019, 19:07:31)
[GCC 7.3.0] linux
10it [00:00, 74367.09it/s]
```
Here is the output in 4.40.0:
```
4.40.0 3.6.9 |Anaconda, Inc.| (default, Jul 30 2019, 19:07:31)
[GCC 7.3.0] linux
0%| | 0/9.6 [00:00<?, ?it/s\
]Traceback (most recent call last):
File "tqdm_test.py", line 3, in <module>
for i in tqdm.tqdm(iterable=range(10), total=9.6):
File "/home/aronnem/miniconda3/envs/tqdm_test/lib/python3.6/site-packages/tqdm/std.py", line 1150, in __iter__
self.close()
File "/home/aronnem/miniconda3/envs/tqdm_test/lib/python3.6/site-packages/tqdm/std.py", line 1261, in close
self.display(pos=0)
File "/home/aronnem/miniconda3/envs/tqdm_test/lib/python3.6/site-packages/tqdm/std.py", line 1428, in display
self.sp(self.__repr__() if msg is None else msg)
File "/home/aronnem/miniconda3/envs/tqdm_test/lib/python3.6/site-packages/tqdm/std.py", line 1058, in __repr__
return self.format_meter(**self.format_dict)
File "/home/aronnem/miniconda3/envs/tqdm_test/lib/python3.6/site-packages/tqdm/std.py", line 482, in format_meter
charset=Bar.ASCII if ascii is True else ascii or Bar.UTF)
File "/home/aronnem/miniconda3/envs/tqdm_test/lib/python3.6/site-packages/tqdm/std.py", line 146, in __init__
assert 0 <= frac <= 1
AssertionError
```
[source website]: https://github.com/tqdm/tqdm/
[known issues]: https://github.com/tqdm/tqdm/#faq-and-known-issues
[issue tracker]: https://github.com/tqdm/tqdm/issues?q=
[StackOverflow#tqdm]: https://stackoverflow.com/questions/tagged/tqdm
|
closed
|
2019-12-05T16:49:54Z
|
2019-12-06T11:29:57Z
|
https://github.com/tqdm/tqdm/issues/859
|
[
"p0-bug-critical ☢",
"duplicate 🗐"
] |
aronnem
| 2
|
mwaskom/seaborn
|
data-visualization
| 3,784
|
[Feature Request] style parameter in `displot`, `catplot` and `lmplot` similar to `relplot`
|
Currently, `relplot` has `style` parameter that provides an additional way to "facet" the data beyond col, row and hue using `linestyle`. It would be nice if this was extended to the other figure plot types. This would also lead to a more consistent API across the different facet grid plots.
- `displot` - kdeplot and ecdfplot would change `linestyle`, histplot would change patch `hatching`.
- `catplot` - stripplot, swarmplot would change `linestyle`; boxplot, violinplot, boxenplot, barplot and countplot would change `hatching`, pointplot would change `linestyle` and `marker`
- `lmplot` - would change `marker` and `linestyle`
References for supporting in underlying matplotlib.
https://matplotlib.org/stable/gallery/lines_bars_and_markers/linestyles.html
https://matplotlib.org/stable/gallery/lines_bars_and_markers/marker_reference.html
https://matplotlib.org/stable/gallery/shapes_and_collections/hatch_style_reference.html
|
closed
|
2024-11-13T21:48:27Z
|
2024-11-13T22:56:40Z
|
https://github.com/mwaskom/seaborn/issues/3784
|
[] |
hguturu
| 4
|
fastapi/fastapi
|
python
| 13,067
|
poor quality traceback / useful stack frames not present when exceptions raised in sync dependencies
|
### Privileged issue
- [X] I'm ~@tiangolo or he asked me directly to create an issue here~ a liar, but the discussion template was crazy onerous, and I'm confident I can write a decent, succinct issue description that's worth reading here ;-)
### Issue Content
If I apply this diff to the [full-stack-fastapi-template](https://github.com/fastapi/full-stack-fastapi-template/blob/88c83cc06ccab67efa839c2c0994435b727986a3/backend/app/api/deps.py#L21-L23):
```diff
diff --git a/backend/app/api/deps.py b/backend/app/api/deps.py
index c2b83c8..c99cdb2 100644
--- a/backend/app/api/deps.py
+++ b/backend/app/api/deps.py
@@ -19,6 +19,7 @@ reusable_oauth2 = OAuth2PasswordBearer(
def get_db() -> Generator[Session, None, None]:
+ raise Exception("error")
with Session(engine) as session:
yield session
```
...and issue a GET to `http://127.0.0.1:8000/api/v1/users/`, the traceback shown in the console is really unhelpful:
```
File "...3.11/lib/python3.11/contextlib.py", line 650, in enter_async_context
result = await _enter(cm)
^^^^^^^^^^^^^^^^
File "...3.11/lib/python3.11/contextlib.py", line 210, in __aenter__
return await anext(self.gen)
^^^^^^^^^^^^^^^^^^^^^
File ".../full-stack-fastapi-template/backend/.venv/lib/python3.11/site-packages/fastapi/concurrency.py", line 35, in contextmanager_in_threadpool
raise e
Exception: error
```
There are no frames from the actual site of the exception, and, on the face of it, potentially no way to go back from the exception to the source of the error.
I first noticed this doing test driven development on a new FastAPI project, having not done any FastAPI dev for a couple of years, and was pretty shocked.
What I don't understand is why the exception being re-raised appears to have no traceback of its own?
|
closed
|
2024-12-13T08:04:35Z
|
2024-12-14T10:42:04Z
|
https://github.com/fastapi/fastapi/issues/13067
|
[] |
cjw296
| 7
|
deepfakes/faceswap
|
machine-learning
| 1,085
|
Being informed on manual preview refresh
|
On slower hardware and with demanding model configurations it can take several minutes until a manual preview refresh actually completes.
For that reason I suggest that another message "Refresh preview done" will be added, so that the user can focus on other things in the meantime and still reliably tell whether the refresh has completed or not.
|
closed
|
2020-11-10T10:37:25Z
|
2021-05-30T10:48:41Z
|
https://github.com/deepfakes/faceswap/issues/1085
|
[
"feature"
] |
OreSeq
| 1
|
rasbt/watermark
|
jupyter
| 1
|
Error in the date with the options -u -n -t -z
|
Hello Sebastian,
I noticed that with `%watermark -u -n -t -z` the date is incorrect, the day takes the same value as the minute. Here is, for comparison, the outputs I have with different options:
---
- Output of %watermark
01/10/2014 14:17:34
CPython 2.7.3
IPython 2.2.0
compiler : GCC 4.2.1 (Apple Inc. build 5666) (dot 3)
system : Darwin
release : 12.5.0
machine : x86_64
processor : i386
CPU cores : 2
interpreter: 64bit
---
- Output of %watermark -d -t
01/10/2014 14:17:34
---
- Output of %watermark -u -n -t -z
Last updated: Wed Oct 17 2014 14:17:34 CEST
|
closed
|
2014-10-01T12:21:08Z
|
2014-10-01T17:46:57Z
|
https://github.com/rasbt/watermark/issues/1
|
[] |
TaurusOlson
| 1
|
521xueweihan/HelloGitHub
|
python
| 2,187
|
微信聊天记录年度报告
|
## 项目推荐
- 项目地址:https://github.com/myth984/wechat-report
- 类别:JS
- 项目后续更新计划:
- 项目描述:
- 必写:和女朋友微信聊天记录统计年度报告
- 描述长度(不包含示例代码): 一步一步的教你生成和女朋友的微信聊天记录年度报告
- 推荐理由:各个APP都有年度报告 给女朋友也出一个报告吧
- 截图:

|
closed
|
2022-04-29T07:51:20Z
|
2022-05-27T01:22:32Z
|
https://github.com/521xueweihan/HelloGitHub/issues/2187
|
[
"已发布",
"JavaScript 项目"
] |
myth984
| 1
|
mwaskom/seaborn
|
data-science
| 3,592
|
Add more detailed errorbar type (ci, pi, se, sd) description to the documentation
|
I propose adding a short explanation of the options ci, pi, sd, and se to every function description that uses the errorbar keyword.
Like a link to the tutorial section for example, since that page never shows up in my search queries.
Showing the equations used behind the different types woul also be a benefit, as it might be unclear if the SDEV is calculated with n or n-1.
|
closed
|
2023-12-14T13:36:39Z
|
2023-12-17T22:43:51Z
|
https://github.com/mwaskom/seaborn/issues/3592
|
[] |
rk-exxec
| 1
|
microsoft/unilm
|
nlp
| 882
|
BEIT v3 code
|
I am very interested in the BEIT v3 paper. When will the code be publicly available? Thanks!
|
closed
|
2022-09-28T12:24:09Z
|
2023-03-13T13:58:55Z
|
https://github.com/microsoft/unilm/issues/882
|
[] |
DecstionBack
| 2
|
babysor/MockingBird
|
deep-learning
| 128
|
RuntimeError: CUDA out of memory. Tried to allocate 58.00 MiB (GPU 0; 6.00 GiB total capacity; 1.83 GiB already allocated; 2.49 GiB free; 2.02 GiB reserved in total by PyTorch)
|
{| Epoch: 1/1 (121/15311) | Loss: 1.719 | 0.73 steps/s | Step: 0k | }Traceback (most recent call last):
出现错误
RuntimeError: CUDA out of memory. Tried to allocate 58.00 MiB (GPU 0; 6.00 GiB total capacity; 1.83 GiB already allocated; 2.49 GiB free; 2.02 GiB reserved in total by PyTorch)
显示显存不足
已经修改### Tacotron Training
tts_schedule = [(2, 1e-3, 10_000, 8), # Progressive training schedule
(2, 5e-4, 15_000, 8), # (r, lr, step, batch_size)
(2, 2e-4, 20_000, 8), # (r, lr, step, batch_size)
(2, 1e-4, 30_000, 8), #
(2, 5e-5, 40_000, 8), #
(2, 1e-5, 60_000, 8), #
(2, 5e-6, 160_000, 8), # r = reduction factor (# of mel frames
(2, 3e-6, 320_000, 8), # synthesized for each decoder iteration)
(2, 1e-6, 640_000, 8)], # lr = learning rate
请问我要怎么调整才能继续训练合成器?感谢各位大佬
|
open
|
2021-10-08T14:47:59Z
|
2022-01-18T12:00:47Z
|
https://github.com/babysor/MockingBird/issues/128
|
[] |
yinjia823
| 4
|
Anjok07/ultimatevocalremovergui
|
pytorch
| 1,577
|
I have this error code, It wont go away, no matter what i try
|
Last Error Received:
Process: MDX-Net
Missing file error raised. Please address the error and try again.
If this error persists, please contact the developers with the error details.
Raw Error Details:
FileNotFoundError: "[WinError 2] Das System kann die angegebene Datei nicht finden"
Traceback Error: "
File "UVR.py", line 6638, in process_start
File "separate.py", line 717, in seperate
File "separate.py", line 382, in final_process
File "separate.py", line 446, in write_audio
File "separate.py", line 419, in save_with_message
File "separate.py", line 393, in save_audio_file
File "separate.py", line 1317, in save_format
File "pydub\audio_segment.py", line 820, in from_wav
File "pydub\audio_segment.py", line 735, in from_file
File "pydub\utils.py", line 274, in mediainfo_json
File "subprocess.py", line 951, in __init__
File "subprocess.py", line 1420, in _execute_child
"
Error Time Stamp [2024-10-05 10:40:26]
Full Application Settings:
vr_model: Choose Model
aggression_setting: 5
window_size: 512
mdx_segment_size: 64
batch_size: Default
crop_size: 256
is_tta: False
is_output_image: False
is_post_process: False
is_high_end_process: False
post_process_threshold: 0.2
vr_voc_inst_secondary_model: No Model Selected
vr_other_secondary_model: No Model Selected
vr_bass_secondary_model: No Model Selected
vr_drums_secondary_model: No Model Selected
vr_is_secondary_model_activate: False
vr_voc_inst_secondary_model_scale: 0.9
vr_other_secondary_model_scale: 0.7
vr_bass_secondary_model_scale: 0.5
vr_drums_secondary_model_scale: 0.5
demucs_model: Choose Model
segment: Default
overlap: 0.25
overlap_mdx: Default
overlap_mdx23: 8
shifts: 2
chunks_demucs: Auto
margin_demucs: 44100
is_chunk_demucs: False
is_chunk_mdxnet: False
is_primary_stem_only_Demucs: False
is_secondary_stem_only_Demucs: False
is_split_mode: True
is_demucs_combine_stems: True
is_mdx23_combine_stems: True
demucs_voc_inst_secondary_model: No Model Selected
demucs_other_secondary_model: No Model Selected
demucs_bass_secondary_model: No Model Selected
demucs_drums_secondary_model: No Model Selected
demucs_is_secondary_model_activate: False
demucs_voc_inst_secondary_model_scale: 0.9
demucs_other_secondary_model_scale: 0.7
demucs_bass_secondary_model_scale: 0.5
demucs_drums_secondary_model_scale: 0.5
demucs_pre_proc_model: No Model Selected
is_demucs_pre_proc_model_activate: False
is_demucs_pre_proc_model_inst_mix: False
mdx_net_model: MDX23C-InstVoc HQ
chunks: Auto
margin: 44100
compensate: Auto
denoise_option: None
is_match_frequency_pitch: True
phase_option: Automatic
phase_shifts: None
is_save_align: False
is_match_silence: True
is_spec_match: False
is_mdx_c_seg_def: False
is_invert_spec: False
is_deverb_vocals: False
deverb_vocal_opt: Main Vocals Only
voc_split_save_opt: Lead Only
is_mixer_mode: False
mdx_batch_size: Default
mdx_voc_inst_secondary_model: No Model Selected
mdx_other_secondary_model: No Model Selected
mdx_bass_secondary_model: No Model Selected
mdx_drums_secondary_model: No Model Selected
mdx_is_secondary_model_activate: False
mdx_voc_inst_secondary_model_scale: 0.9
mdx_other_secondary_model_scale: 0.7
mdx_bass_secondary_model_scale: 0.5
mdx_drums_secondary_model_scale: 0.5
is_save_all_outputs_ensemble: True
is_append_ensemble_name: False
chosen_audio_tool: Manual Ensemble
choose_algorithm: Min Spec
time_stretch_rate: 2.0
pitch_rate: 2.0
is_time_correction: True
is_gpu_conversion: True
is_primary_stem_only: False
is_secondary_stem_only: False
is_testing_audio: False
is_auto_update_model_params: True
is_add_model_name: False
is_accept_any_input: False
is_task_complete: False
is_normalization: False
is_use_opencl: True
is_wav_ensemble: False
is_create_model_folder: False
mp3_bit_set: 320k
semitone_shift: 0
save_format: FLAC
wav_type_set: 64-bit Float
device_set: NVIDIA GeForce RTX 3070:0
help_hints_var: True
set_vocal_splitter: No Model Selected
is_set_vocal_splitter: False
is_save_inst_set_vocal_splitter: False
model_sample_mode: False
model_sample_mode_duration: 30
demucs_stems: All Stems
mdx_stems: Vocals
|
open
|
2024-10-05T08:41:42Z
|
2024-10-05T08:41:42Z
|
https://github.com/Anjok07/ultimatevocalremovergui/issues/1577
|
[] |
Elias02345
| 0
|
facebookresearch/fairseq
|
pytorch
| 4,633
|
Transformer's pad token has non-zero embedding
|
## 🐛 Bug
By default a transformer's token embedding layer has `padding_idx=1` indicating that the embedding of this token should be a 0 vector and should not contribute to loss. However, transformers trained using fairseq have non-zero pad token embeddings.
### To Reproduce
Steps to reproduce the behavior (**always include the command you ran**):
1. Train a model
2. Look at the token embedding weights
#### Code sample
The following fails with a transformer trained using fairseq:
```python
def check_zero_embedding(checkpoint_path, padding_idx=1):
weights = torch.load(checkpoint_path)['model']['encoder.embed_tokens.weight']
assert torch.all(weights[padding_idx] == 0)
```
alternatively, examine for WMT19 system:
```python
#!/usr/bin/env python3
import torch
import logging
# disable noisy fairseq logging...
logging.disable(1_000)
if __name__ == "__main__":
en2de = torch.hub.load('pytorch/fairseq', 'transformer.wmt19.en-de.single_model')
pad_embedding_weight = en2de.models[0].encoder.embed_tokens.weight[en2de.models[0].encoder.embed_tokens.padding_idx]
assert torch.all(pad_embedding_weight == 0), f"Embedding for padding index should be 0, found {pad_embedding_weight}"
```
which fails:
```console
$ python failing_test.py
Using cache found in /Users/erip/.cache/torch/hub/pytorch_fairseq_main
Loading codes from /Users/erip/.cache/torch/pytorch_fairseq/81a0be5cbbf1c106320ef94681844d4594031c94c16b0475be11faa5a5120c48.63b093d59e7e0814ff799bb965ed4cbde30200b8c93a44bf8c1e5e98f5c54db3/bpecodes ...
Read 30000 codes from the codes file.
Traceback (most recent call last):
File "/Users/erip/failing_test.py", line 11, in <module>
assert torch.all(pad_embedding_weight == 0), f"Embedding for padding index should be 0, found {pad_embedding_weight}"
AssertionError: Embedding for padding index should be 0, found tensor([ 0.0109, 0.0018, 0.0024, ..., 0.0170, -0.0071, -0.0250],
grad_fn=<SelectBackward0>)
```
### Expected behavior
<!-- A clear and concise description of what you expected to happen. -->
### Environment
- fairseq Version (e.g., 1.0 or main): main
- PyTorch Version (e.g., 1.0) 1.12
- OS (e.g., Linux): n/a
- How you installed fairseq (`pip`, source): source
- Build command you used (if compiling from source): ...
- Python version:
- CUDA/cuDNN version:
- GPU models and configuration:
- Any other relevant information:
### Additional context
<!-- Add any other context about the problem here. -->
|
open
|
2022-08-07T00:03:59Z
|
2022-08-09T13:53:12Z
|
https://github.com/facebookresearch/fairseq/issues/4633
|
[
"bug",
"needs triage"
] |
erip
| 3
|
lux-org/lux
|
jupyter
| 94
|
Improve general sampling strategy in PandasExecutor
|
The current sampling strategy is crude and largely based on random sampling. We should investigate the performance degradation of Lux across various large datasets to select better sampling strategies, as well as exposing tunable parameters in the API to allow users to adjust for different sampling parameters and strategies. Add ReadTheDoc page explaining default sampling strategy and how to tune them in Lux.
|
closed
|
2020-09-25T08:49:56Z
|
2021-01-08T10:03:58Z
|
https://github.com/lux-org/lux/issues/94
|
[] |
dorisjlee
| 1
|
gevent/gevent
|
asyncio
| 1,777
|
SSLContext recursion when initialized before patching
|
Simple repro:
```python
import gevent.monkey
import ssl
ctx = ssl.SSLContext()
ctx.options |= ssl.Options.OP_NO_TICKET
gevent.monkey.patch_all()
ctx.options |= ssl.Options.OP_NO_TICKET # infinite recursion here
```
Perhaps a way to solve it would be to restructure `gevent._ssl3.SSLContext` to **copy** SSLContext instead of **basing** on it:
```diff
-class SSLContext(orig_SSLContext):
+class SSLContext(orig_SSLContext.__base__):
__slots__ = ()
+ vars().update(orig_SSLContext.__dict__)
```
|
closed
|
2021-03-03T20:01:21Z
|
2021-03-04T02:03:24Z
|
https://github.com/gevent/gevent/issues/1777
|
[] |
ikonst
| 4
|
google-research/bert
|
tensorflow
| 706
|
Couldn't train BERT with SQUAD 1.1
|
I created VM (n1-standard-2) and Cloud TPU (v3-8) using ctpu tool.
I have created a Storage bucket and mounted it in VM using GCSfuse.
Tried to run it. Failed.
```
python run_squad.py \
> --vocab_file=$BERT_BASE_DIR/vocab.txt \
> --bert_config_file=$BERT_BASE_DIR/bert_config.json \
> --init_checkpoint=$BERT_BASE_DIR/bert_model.ckpt \
> --do_train=True \
> --train_file=$SQUAD_DIR/train-v1.1.json \
> --do_predict=True \
> --predict_file=$SQUAD_DIR/dev-v1.1.json \
> --train_batch_size=12 \
> --learning_rate=3e-5 \
> --num_train_epochs=2.0 \
> --max_seq_length=384 \
> --doc_stride=128 \
> output_dir=$output
Traceback (most recent call last):
File "run_squad.py", line 1283, in <module>
tf.app.run()
File "/usr/local/lib/python2.7/dist-packages/tensorflow/python/platform/app.py", l
ine 119, in run
argv = flags.FLAGS(_sys.argv if argv is None else argv, known_only=True)
File "/usr/local/lib/python2.7/dist-packages/tensorflow/python/platform/flags.py",
line 112, in __call__
return self.__dict__['__wrapped'].__call__(*args, **kwargs)
File "/usr/local/lib/python2.7/dist-packages/absl/flags/_flagvalues.py", line 636,
in __call__
self._assert_all_validators()
File "/usr/local/lib/python2.7/dist-packages/absl/flags/_flagvalues.py", line 510,
in _assert_all_validators
self._assert_validators(all_validators)
File "/usr/local/lib/python2.7/dist-packages/absl/flags/_flagvalues.py", line 531,
in _assert_validators
raise _exceptions.IllegalFlagValueError('%s: %s' % (message, str(e)))
absl.flags._exceptions.IllegalFlagValueError: flag --output_dir=None: Flag --output_
dir must have a value other than None.
```
`$output` is `/output: Is a directory`.
|
closed
|
2019-06-19T06:13:50Z
|
2023-02-17T04:17:26Z
|
https://github.com/google-research/bert/issues/706
|
[] |
JeevaTM
| 4
|
mlfoundations/open_clip
|
computer-vision
| 261
|
Investigate memory usage for HF models
|
mt5 xl non frozen + H/14 frozen uses > 40GB at batch size 1
that seems wrong, something is probably off
|
closed
|
2022-11-28T01:39:05Z
|
2024-10-30T16:32:06Z
|
https://github.com/mlfoundations/open_clip/issues/261
|
[
"bug",
"important"
] |
rom1504
| 2
|
vitalik/django-ninja
|
rest-api
| 436
|
Make `NinjaAPI().add_router` idempotent
|
**Problem**
I have a project where we attach/register routes/urls during `apps.ready`. Which becomes problematic to keep doing when using `django-ninja` while also having test cases calling
```python
with django.test.utils.modify_settings(INSTALLED_APPS={"append": ["django-app"]}):
...
```
As that'll trigger a runthrough of `apps.ready` for each installed app and `NinjaAPI().add_router` isn't idempotent, but instead raises a `ConfigError` from the code below:
https://github.com/vitalik/django-ninja/blob/22e97cdab9faabc84a048eeac688192f3f1f19d7/ninja/router.py#L352-L359
**Describe the solution you'd like**
I'd like `NinjaAPI().add_router` to be idempotent. Which I, from my brief investigation, think is possible. Since a `NinjaAPI` instance stores a list of 2-tuples with each tuple containing a prefix and a router. So what if `NinjaAPI.add_router` starts out by checking
```python
if router.api == self and (prefix, router) in self._routers:
# Idempotent on identical registration
return
elif router.api is not None:
# Router already attached to an api
raise ConfigError(...)
```
I think that should also allow removal of the `debug_server_url_reimport` in `Router.build_routers`:
https://github.com/vitalik/django-ninja/blob/22e97cdab9faabc84a048eeac688192f3f1f19d7/ninja/router.py#L354-L356
|
open
|
2022-05-04T07:50:31Z
|
2022-05-04T07:50:31Z
|
https://github.com/vitalik/django-ninja/issues/436
|
[] |
flaeppe
| 0
|
RobertCraigie/prisma-client-py
|
asyncio
| 259
|
Create with required relation is incorrectly typed
|
<!--
Thanks for helping us improve Prisma Client Python! 🙏 Please follow the sections in the template and provide as much information as possible about your problem, e.g. by enabling additional logging output.
See https://prisma-client-py.readthedocs.io/en/stable/reference/logging/ for how to enable additional logging output.
-->
## Bug description
<!-- A clear and concise description of what the bug is. -->
The following code will pass type checks but will raise an error at runtime as user is a required relation
```py
Profile.prisma().create({'bio': 'My bio', 'country': 'Scotland'})
```
## Expected behavior
<!-- A clear and concise description of what you expected to happen. -->
Type checkers should report an error
## Prisma information
<!-- Your Prisma schema, Prisma Client Python queries, ...
Do not include your database credentials when sharing your Prisma schema! -->
Internal schema
## Environment & setup
<!-- In which environment does the problem occur -->
- OS: <!--[e.g. Mac OS, Windows, Debian, CentOS, ...]--> Mac OS
- Database: <!--[PostgreSQL, MySQL, MariaDB or SQLite]--> SQLite
- Python version: <!--[Run `python -V` to see your Python version]--> 3.9.9
|
open
|
2022-02-01T01:16:11Z
|
2022-02-02T03:11:40Z
|
https://github.com/RobertCraigie/prisma-client-py/issues/259
|
[
"bug/2-confirmed",
"kind/bug",
"level/intermediate",
"priority/high"
] |
RobertCraigie
| 0
|
recommenders-team/recommenders
|
data-science
| 1,597
|
[BUG] Error when publishing to pypi with pymanopt
|
### Description
<!--- Describe your issue/bug/request in detail -->
When publishing to pypi the current code, I get an error:
```
$ twine upload lib/*
Uploading distributions to https://upload.pypi.org/legacy/
/anaconda/envs/py38_default/lib/python3.8/site-packages/twine/auth.py:66: UserWarning: Failed to create the collection: Prompt dismissed..
warnings.warn(str(exc))
Enter your username: miguelgfierro
/anaconda/envs/py38_default/lib/python3.8/site-packages/twine/auth.py:75: UserWarning: Failed to create the collection: Prompt dismissed..
warnings.warn(str(exc))
Enter your password:
Uploading recommenders-1.0.0-py3-none-manylinux1_x86_64.whl
100%|██████████████████████████████████████████████████████████████████████████████████████████████████| 334k/334k [00:01<00:00, 309kB/s]
Error during upload. Retry with the --verbose option for more details.
HTTPError: 400 Bad Request from https://upload.pypi.org/legacy/
Invalid value for requires_dist. Error: Can't have direct dependency: 'pymanopt @ https://github.com/pymanopt/pymanopt/archive/fb36a272cdeecb21992cfd9271eb82baafeb316d.zip'
```
We need to find a good dependency for pymanopt that works with TF2 and the numpy version we have
### In which platform does it happen?
<!--- Describe the platform where the issue is happening (use a list if needed) -->
<!--- For example: -->
<!--- * Azure Data Science Virtual Machine. -->
<!--- * Azure Databricks. -->
<!--- * Other platforms. -->
### How do we replicate the issue?
<!--- Please be specific as possible (use a list if needed). -->
<!--- For example: -->
<!--- * Create a conda environment for pyspark -->
<!--- * Run unit test `test_sar_pyspark.py` with `pytest -m 'spark'` -->
<!--- * ... -->
### Expected behavior (i.e. solution)
<!--- For example: -->
<!--- * The tests for SAR PySpark should pass successfully. -->
### Other Comments
|
closed
|
2021-12-21T13:26:08Z
|
2022-01-10T14:50:40Z
|
https://github.com/recommenders-team/recommenders/issues/1597
|
[
"bug"
] |
miguelgfierro
| 7
|
syrupy-project/syrupy
|
pytest
| 438
|
How to use pytest.approx() with syrupy
|
We have a bunch of tests that perform computations using pandas, numpy, and other scientific libraries and produce a dictionary containing the resulting key-value pairs. Some of the values are slightly different when run on different platforms (i.e. macOS vs Linux), so we wrap those values with `pytest.approx()` to accommodate those minor and acceptable differences.
Digging into the syrupy code, the final comparison between the test value and the snapshot is performed against serialized data, so it appears that `pytest.approx()` cannot be used. Is that correct? Or can you suggest a way to allow these two great features to be used together?
Thanks!
|
closed
|
2020-12-01T00:30:51Z
|
2020-12-03T17:30:22Z
|
https://github.com/syrupy-project/syrupy/issues/438
|
[
"question"
] |
hashstat
| 5
|
Textualize/rich
|
python
| 3,441
|
[REQUEST] Support python -m rich.traceback myscript.py
|
**How would you improve Rich?**
The Python debugger `pdb` supports running a script with debugging enabled by changing the command line call from `python myscript.py --args` to `python -m pdb myscript.py --args`. It would be great to have the same functionality for running a script with rich tracebacks installed: `python -m rich.traceback myscript.py --args`. The argument-less `python -m rich.traceback` could continue to display an example as before.
**What problem does it solve for you?**
This feature would allow using rich tracebacks occasionally for debugging without requiring to change any code or installing `sitecustomize.py` (especially when frequently changing environments).
|
closed
|
2024-08-02T05:59:18Z
|
2024-08-26T14:58:35Z
|
https://github.com/Textualize/rich/issues/3441
|
[
"Needs triage"
] |
f0k
| 3
|
Guovin/iptv-api
|
api
| 640
|
分类指的是?这个地址作为订阅源我试了是可以获取结果的
|
分类指的是?这个地址作为订阅源我试了是可以获取结果的
_Originally posted by @Guovin in https://github.com/Guovin/iptv-api/issues/637#issuecomment-2530116885_
是可以在每个大类上细分,如咪咕分类下有CCTV1-17,卫视等等,还有IPV6分类下有CCTV1-17,卫视等等。有没可能按照链接上的来分类
|
closed
|
2024-12-10T07:40:13Z
|
2024-12-10T07:41:34Z
|
https://github.com/Guovin/iptv-api/issues/640
|
[] |
alantang1977
| 0
|
ml-tooling/opyrator
|
pydantic
| 94
|
Issue regarding determine uploaded file types on MIME
|
Hi, i played a bit with the project and noticed one potential issue. In this [function](https://github.com/ml-tooling/opyrator/blob/3f443f05b6b21f00685c2b9bba16cf080edf2385/src/opyrator/ui/streamlit_ui.py#L242), the mime type could be manipulated by remote user, hence he could upload any file with a manipulated MIME header. The description of such potential vulnerability is [here](https://owasp.org/www-community/vulnerabilities/Unrestricted_File_Upload#Using_White-List_for_Files.E2.80.99_Extensions). One could use magic code to check the uploaded file type rather than rely on the MIME or extension
|
closed
|
2024-02-18T20:04:21Z
|
2024-11-08T02:33:18Z
|
https://github.com/ml-tooling/opyrator/issues/94
|
[
"question",
"stale"
] |
nevercodecorrect
| 3
|
microsoft/Bringing-Old-Photos-Back-to-Life
|
pytorch
| 166
|
老照片
|
老照片
|
closed
|
2021-05-15T16:23:15Z
|
2021-05-24T11:02:35Z
|
https://github.com/microsoft/Bringing-Old-Photos-Back-to-Life/issues/166
|
[] |
56518
| 0
|
pyro-ppl/numpyro
|
numpy
| 1,916
|
`MCMC.run` gets error after `MCMC.warmup` with `AIES`
|
Hi, I get an error when I `MCMC.run` after `warmup` with `AIES`.
Here is the example
```
import jax
import jax.numpy as jnp
import numpyro
from numpyro.infer import MCMC, AIES
import numpyro.distributions as dist
n_dim, num_chains = 5, 100
mu, sigma = jnp.zeros(n_dim), jnp.ones(n_dim)
def model(mu, sigma):
with numpyro.plate('n_dim', n_dim):
numpyro.sample("x", dist.Normal(mu, sigma))
kernel = AIES(model, moves={AIES.DEMove() : 0.5,
AIES.StretchMove() : 0.5})
mcmc = MCMC(kernel,
num_warmup=100,
num_samples=100,
num_chains=num_chains,
chain_method='vectorized')
mcmc.warmup(jax.random.PRNGKey(0), mu, sigma)
mcmc.run(jax.random.PRNGKey(1), mu, sigma)
```
The error
```
{
"name": "ValueError",
"message": "split accepts a single key, but was given a key array of shape (100, 2) != (). Use jax.vmap for batching.",
"stack": "---------------------------------------------------------------------------
ValueError Traceback (most recent call last)
Cell In[13], line 26
18 mcmc = MCMC(kernel,
19 num_warmup=100,
20 num_samples=100,
21 num_chains=num_chains,
22 chain_method='vectorized')
25 mcmc.warmup(jax.random.PRNGKey(0), mu, sigma)
---> 26 mcmc.run(jax.random.PRNGKey(1), mu, sigma)
File ~/anaconda3/lib/python3.11/site-packages/numpyro/infer/mcmc.py:675, in MCMC.run(self, rng_key, extra_fields, init_params, *args, **kwargs)
673 else:
674 assert self.chain_method == \"vectorized\"
--> 675 states, last_state = partial_map_fn(map_args)
676 # swap num_samples x num_chains to num_chains x num_samples
677 states = tree_map(lambda x: jnp.swapaxes(x, 0, 1), states)
File ~/anaconda3/lib/python3.11/site-packages/numpyro/infer/mcmc.py:462, in MCMC._single_chain_mcmc(self, init, args, kwargs, collect_fields, remove_sites)
456 collection_size = self._collection_params[\"collection_size\"]
457 collection_size = (
458 collection_size
459 if collection_size is None
460 else collection_size // self.thinning
461 )
--> 462 collect_vals = fori_collect(
463 lower_idx,
464 upper_idx,
465 sample_fn,
466 init_val,
467 transform=_collect_fn(collect_fields, remove_sites),
468 progbar=self.progress_bar,
469 return_last_val=True,
470 thinning=self.thinning,
471 collection_size=collection_size,
472 progbar_desc=partial(_get_progbar_desc_str, lower_idx, phase),
473 diagnostics_fn=diagnostics,
474 num_chains=self.num_chains if self.chain_method == \"parallel\" else 1,
475 )
476 states, last_val = collect_vals
477 # Get first argument of type `HMCState`
File ~/anaconda3/lib/python3.11/site-packages/numpyro/util.py:367, in fori_collect(lower, upper, body_fun, init_val, transform, progbar, return_last_val, collection_size, thinning, **progbar_opts)
365 with tqdm.trange(upper) as t:
366 for i in t:
--> 367 vals = jit(_body_fn)(i, vals)
368 t.set_description(progbar_desc(i), refresh=False)
369 if diagnostics_fn:
[... skipping hidden 11 frame]
File ~/anaconda3/lib/python3.11/site-packages/numpyro/util.py:332, in fori_collect.<locals>._body_fn(i, vals)
329 @cached_by(fori_collect, body_fun, transform)
330 def _body_fn(i, vals):
331 val, collection, start_idx, thinning = vals
--> 332 val = body_fun(val)
333 idx = (i - start_idx) // thinning
334 collection = cond(
335 idx >= 0,
336 collection,
(...)
339 identity,
340 )
File ~/anaconda3/lib/python3.11/site-packages/numpyro/infer/mcmc.py:188, in _sample_fn_nojit_args(state, sampler, args, kwargs)
186 def _sample_fn_nojit_args(state, sampler, args, kwargs):
187 # state is a tuple of size 1 - containing HMCState
--> 188 return (sampler.sample(state[0], args, kwargs),)
File ~/anaconda3/lib/python3.11/site-packages/numpyro/infer/ensemble.py:192, in EnsembleSampler.sample(self, state, model_args, model_kwargs)
190 def sample(self, state, model_args, model_kwargs):
191 z, inner_state, rng_key = state
--> 192 rng_key, _ = random.split(rng_key)
193 z_flat, unravel_fn = batch_ravel_pytree(z)
195 if self._randomize_split:
File ~/anaconda3/lib/python3.11/site-packages/jax/_src/random.py:285, in split(key, num)
274 def split(key: KeyArrayLike, num: int | tuple[int, ...] = 2) -> KeyArray:
275 \"\"\"Splits a PRNG key into `num` new keys by adding a leading axis.
276
277 Args:
(...)
283 An array-like object of `num` new PRNG keys.
284 \"\"\"
--> 285 typed_key, wrapped = _check_prng_key(\"split\", key, error_on_batched=True)
286 return _return_prng_keys(wrapped, _split(typed_key, num))
File ~/anaconda3/lib/python3.11/site-packages/jax/_src/random.py:108, in _check_prng_key(name, key, allow_batched, error_on_batched)
105 msg = (f\"{name} accepts a single key, but was given a key array of \"
106 f\"shape {np.shape(key)} != (). Use jax.vmap for batching.\")
107 if error_on_batched:
--> 108 raise ValueError(msg)
109 else:
110 warnings.warn(msg + \" In a future JAX version, this will be an error.\",
111 FutureWarning, stacklevel=3)
ValueError: split accepts a single key, but was given a key array of shape (100, 2) != (). Use jax.vmap for batching."
```
It will get the same error if use `ESS`
|
closed
|
2024-11-25T14:53:29Z
|
2024-11-29T01:52:21Z
|
https://github.com/pyro-ppl/numpyro/issues/1916
|
[
"bug"
] |
xiesl97
| 2
|
aiogram/aiogram
|
asyncio
| 1,103
|
Documentation mistake for PreCheckoutQuery type.
|
### Checklist
- [x] I am sure the error is coming from aiogram code
- [X] I have searched in the issue tracker for similar bug reports, including closed ones
### Operating system
Windows 11 21H2
### Python version
3.11.1
### aiogram version
2.24
### Expected behavior
When I open documentation for [types.PreCheckoutQuery](https://github.com/aiogram/aiogram/blob/dev-2.x/aiogram/types/pre_checkout_query.py) class, it should contain docstring as provided in [Bot API documentation](https://core.telegram.org/bots/api#precheckoutquery).
### Current behavior
When I open documentation for [types.PreCheckoutQuery](https://github.com/aiogram/aiogram/blob/dev-2.x/aiogram/types/pre_checkout_query.py) class, I see text in docstring about HTML5 games which is not related to Telegram Payments and `PreCheckoutQuery` type.
### Steps to reproduce
1. Open [source code](https://github.com/aiogram/aiogram/blob/dev-2.x/aiogram/types/pre_checkout_query.py) for PreCheckoutQuery type.
2. Open official Bot API [documentation](https://core.telegram.org/bots/api#precheckoutquery).
3. Check that docstring in code and description in documentation do not match as they should be.
### Code example
_No response_
### Logs
_No response_
### Additional information
This is also reproduced when I open documentation on [docs.aiogram.dev](https://docs.aiogram.dev/en/latest/telegram/types/pre_checkout_query.html) in the browser
|
closed
|
2023-01-21T12:44:49Z
|
2023-08-14T20:23:56Z
|
https://github.com/aiogram/aiogram/issues/1103
|
[
"bug"
] |
iamnalinor
| 1
|
sinaptik-ai/pandas-ai
|
data-visualization
| 841
|
Conversational/Follow Up questions for Agent still takes the base dataframe for code generation
|
### System Info
python==3.10.13
pandasai==1.5.11
Windows OS
### 🐛 Describe the bug
I initialized pandasai agent for conversation capabilities. Gave the base dataframe and Azure OpenAI LLM. Agent answers first question well, but when I ask follow up question it runs/build the python code on base dataframe rather than previous answer dataframe. Below is my code (excluding imports)
```
nl_course_agent = Agent([course_data], memory_size=10, config={
"llm": llm, "response_parser": CustomPandasDataFrameResponse,
"generate_python_code": MyCustomPrompt()
# "custom_instructions": "Ensure to include all queries in the conversation while you generate the response"
}
)
question1 = "what are HR case management related courses?"
question2 = "show me only greater than 4 rating"
nl_course_agent.start_new_conversation()
nl_course_agent.chat(question1) ## returns right answer
### Follow up questions
nl_course_agent.chat(question2) ## Returns wrong answer
print(nl_course_agent.last_prompt)
<conversation>
Q: what are HR case management related courses?
A: Check it out: <dataframe>
</conversation>
<query>
show me only greater than 4 rating
</query>
Is the query somehow related to the previous conversation? Answer only "true" or "false" (lowercase).
nl_course_agent.check_if_related_to_conversation(question2) ## Returns True
```
I also tried a custom prompt but no change in response.
```
class MyCustomPrompt(AbstractPrompt):
template = """You are given a dataframe with number if rows equal to {dfs[0].shape[0]} and number of columns equal to {dfs[0].shape[1]}
Here's the conversation:
{conversation}
If the question is related to conversation, then use entire conversation to filter the dataframe. Not just the recent question.
"""
```
How to make agent
1) Take entire conversation to build python code if it recognizes a follow up question ? OR
2) Pass filtered dataframe as input to follow up question?
|
closed
|
2023-12-29T05:11:32Z
|
2024-06-01T00:20:42Z
|
https://github.com/sinaptik-ai/pandas-ai/issues/841
|
[] |
chaituValKanO
| 0
|
itamarst/eliot
|
numpy
| 144
|
Allow passing existing `Field`s to `fields`
|
As it stands combining previously defined `Field`s with `fields` is a bit of a mess:
``` python
BAR = Field(u'bar', ...)
LOG_FOO = ActionType(u'example:foo', [BAR, ...] + fields(reason=int, ...), [BAR, ...] + fields(result=int), ...)
```
It would be convenient if `fields` accepted positional arguments of `Field` instances.
|
closed
|
2015-02-24T09:43:19Z
|
2018-09-22T20:59:16Z
|
https://github.com/itamarst/eliot/issues/144
|
[] |
jonathanj
| 1
|
keras-team/autokeras
|
tensorflow
| 828
|
[Question] Project Algorithms Milestones and its relations to Kerastuner
|
Hi,
You seem to be using RandomSearch and params band search (HyperBand) from kerastuner as algorithms. You are planning to deploy other popular NAS/AutoML as Reinforcement Learning and Meta-Heuristic Search (GA e.g.) ? Does keras-team plans to do all the algorithms in kerastuner or in this module ? Do you have a road to the milestones of it until the beta ?
NAS is a very new research topic and seems to be very exciting to build a module to easy the flow through this research ...
To note: master branch is not specifying kerastuner as dependency as-far-i-have-seem.
thank you
|
closed
|
2019-11-07T19:04:30Z
|
2020-02-08T14:39:21Z
|
https://github.com/keras-team/autokeras/issues/828
|
[
"wontfix"
] |
Uiuran
| 6
|
rougier/scientific-visualization-book
|
numpy
| 7
|
Visualizations
|
open
|
2021-08-11T23:06:12Z
|
2021-08-11T23:06:12Z
|
https://github.com/rougier/scientific-visualization-book/issues/7
|
[] |
morr-debug
| 0
|
|
floodsung/Deep-Learning-Papers-Reading-Roadmap
|
deep-learning
| 85
|
UnicodeEncodeError: 'charmap' codec can't encode
|
[3] "Reducing the dimensionality of data with neur [...] (http://www.cs.toronto.edu/~hinton/science.pdf)
Traceback (most recent call last):
File "download.py", line 102, in <module>
print_title(point.text)
File "download.py", line 50, in print_title
print('\n'.join(("", title, pattern * len(title))))
File "C:\Python27\lib\encodings\cp437.py", line 12, in encode
return codecs.charmap_encode(input,errors,encoding_map)
UnicodeEncodeError: 'charmap' codec can't encode character u'\uff08' in position 23: character maps to <undefined>
|
open
|
2017-12-05T15:37:13Z
|
2019-06-10T12:47:44Z
|
https://github.com/floodsung/Deep-Learning-Papers-Reading-Roadmap/issues/85
|
[] |
Vyomax
| 4
|
tensorpack/tensorpack
|
tensorflow
| 782
|
ops are assigned to gpu when run DistributedTrainerParameterServer
|
I want to run DistributedTrainerParameterServer example , so I modify thr examples/basics/mnist-convnet.py as below:
launch_train_with_config(config, SyncMultiGPUTrainerParameterServer(gpus=args.gpu.split(",")))
===>
cluster = tf.train.ClusterSpec({"ps":["localhost:2222"], "worker":["localhost:2223"]})
server = tf.train.Server(cluster,
job_name=args.job_name,
task_index=int(args.task_index))
launch_train_with_config(config, DistributedTrainerParameterServer([int(x) for x in args.gpu.split(",")], server))
and run it in two window:
python distributed-mnist-convnet.py --gpu=0 --job_name=ps --task_index=0
python distributed-mnist-convnet.py --gpu=0 --job_name=worker --task_index=0
then I got the error:
`
[0606 09:37:48 @summary.py:75] Summarizing collection 'summaries' of size 19.
[0606 09:37:48 @base.py:197] Creating the session ...
2018-06-06 09:37:48.721536: I tensorflow/core/distributed_runtime/master_session.cc:1024] Start master session 840fd3c8777697e2 with config:
Traceback (most recent call last):
File "distributed-mnist-convnet.py", line 145, in <module>
launch_train_with_config(config, DistributedTrainerParameterServer([int(x) for x in args.gpu.split(",")], server))
File "/usr/local/lib/python2.7/dist-packages/tensorpack-0.8.5-py2.7.egg/tensorpack/train/interface.py", line 90, in launch_train_with_config
extra_callbacks=config.extra_callbacks)
File "/usr/local/lib/python2.7/dist-packages/tensorpack-0.8.5-py2.7.egg/tensorpack/train/base.py", line 301, in train_with_defaults
steps_per_epoch, starting_epoch, max_epoch)
File "/usr/local/lib/python2.7/dist-packages/tensorpack-0.8.5-py2.7.egg/tensorpack/train/base.py", line 272, in train
self.initialize(session_creator, session_init)
File "/usr/local/lib/python2.7/dist-packages/tensorpack-0.8.5-py2.7.egg/tensorpack/train/trainers.py", line 199, in initialize
get_distributed_session_creator(self.server), session_init)
File "/usr/local/lib/python2.7/dist-packages/tensorpack-0.8.5-py2.7.egg/tensorpack/utils/argtools.py", line 181, in wrapper
return func(*args, **kwargs)
File "/usr/local/lib/python2.7/dist-packages/tensorpack-0.8.5-py2.7.egg/tensorpack/train/base.py", line 200, in initialize
self.sess = session_creator.create_session()
File "/usr/local/lib/python2.7/dist-packages/tensorpack-0.8.5-py2.7.egg/tensorpack/tfutils/distributed.py", line 40, in create_session
return sm.prepare_session(master=server.target, init_op=init_op)
File "/usr/local/lib/python2.7/dist-packages/tensorflow/python/training/session_manager.py", line 281, in prepare_session
sess.run(init_op, feed_dict=init_feed_dict)
File "/usr/local/lib/python2.7/dist-packages/tensorflow/python/client/session.py", line 905, in run
run_metadata_ptr)
File "/usr/local/lib/python2.7/dist-packages/tensorflow/python/client/session.py", line 1140, in _run
feed_dict_tensor, options, run_metadata)
File "/usr/local/lib/python2.7/dist-packages/tensorflow/python/client/session.py", line 1321, in _do_run
run_metadata)
File "/usr/local/lib/python2.7/dist-packages/tensorflow/python/client/session.py", line 1340, in _do_call
raise type(e)(node_def, op, message)
tensorflow.python.framework.errors_impl.InvalidArgumentError: Cannot assign a device for operation 'tower0/lr': Could not satisfy explicit device specification '/job:worker/task:0/device:GPU:0' because no supported kernel for GPU devices is available.
Registered kernels:
device='CPU'; T in [DT_DOUBLE]
device='CPU'; T in [DT_FLOAT]
device='CPU'; T in [DT_BFLOAT16]
device='CPU'; T in [DT_HALF]
device='CPU'; T in [DT_INT8]
device='CPU'; T in [DT_UINT8]
device='CPU'; T in [DT_INT16]
device='CPU'; T in [DT_UINT16]
device='CPU'; T in [DT_INT32]
device='CPU'; T in [DT_INT64]
[[Node: tower0/lr = ScalarSummary[T=DT_FLOAT, _device="/job:worker/task:0/device:GPU:0"](tower0/lr/tags, tower0/learning_rate)]]
`
|
closed
|
2018-06-06T09:47:26Z
|
2018-06-15T07:40:19Z
|
https://github.com/tensorpack/tensorpack/issues/782
|
[
"usage",
"upstream issue"
] |
yogurfrul
| 5
|
DistrictDataLabs/yellowbrick
|
scikit-learn
| 1,087
|
Show ROC for train and test + hide macro/micro in binary classification.
|
Hi! Thanks (again) for this incredible package 👍
I hope not to be missing heavily with this feature, I read all of the documentation + google + stackoverflow. Please if I'm wrong to point me in the right direction (=> URL)
Normally we check train data vs test data. I see that ROC feature only shows the data for test. It would be nice to see the whole picture and check if the model is overfitting.
But it is not easy in multiclass.
In addition, I'd like to know how micro/macro statistics are calculated in binary class. Never see it before (and I guess it is only useful in multiclass). Thus, my recommendation is to turn them False by default.
It can be done smoothly in sklearn:
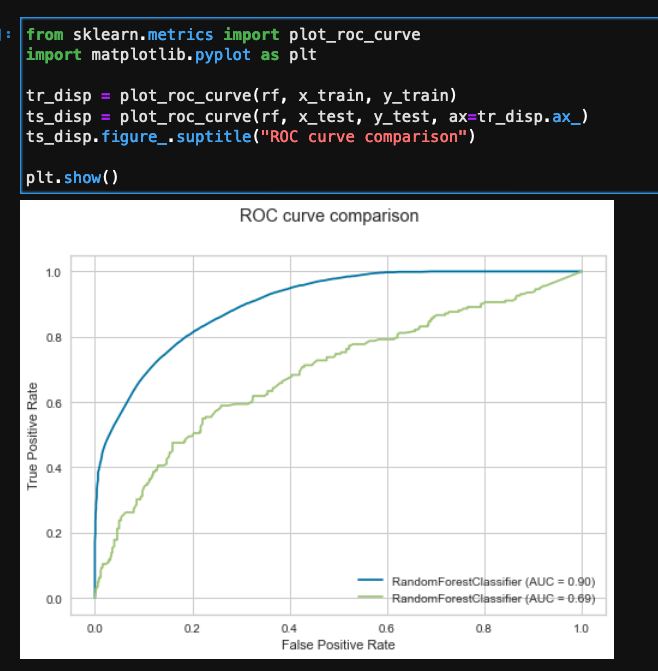
In yellowbrick, for the same data I see:
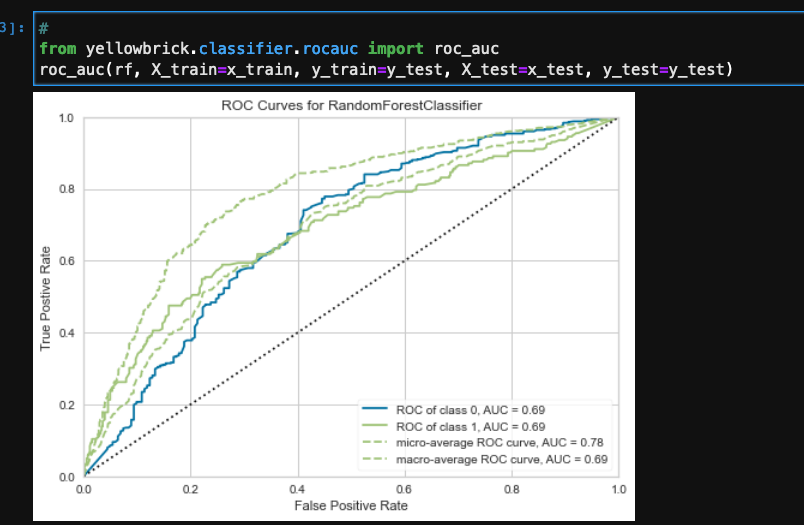
As you can see, in the last plot the information regarding micro/macro does not add value, but the fact that the model heavily overfitting cannot be missed.
This is my hack, to pass the train data as test data as shown in the image:
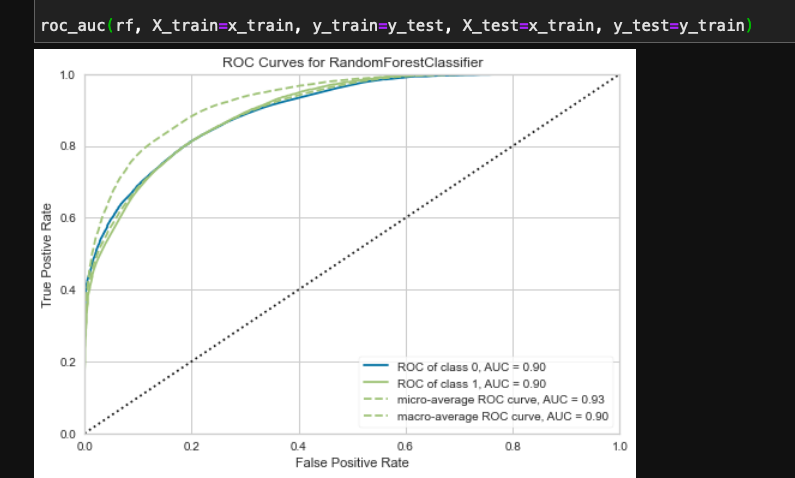
Given the mentioned behavior, why do we have to pass the train in order to plot since the current plot only shows the data for testing? Wouldn't be enough the model + the test data?
Finally, another suggestion is in binary classification, to hide less representative class. It shouldn't add too much value to show both classes.
Thanks a lot in advance! Keep rocking 🤘
|
closed
|
2020-07-18T23:39:51Z
|
2020-10-02T17:43:27Z
|
https://github.com/DistrictDataLabs/yellowbrick/issues/1087
|
[
"type: question"
] |
pablo14
| 4
|
timkpaine/lantern
|
plotly
| 201
|
Add conda recipe
|
closed
|
2020-02-18T19:59:13Z
|
2024-02-03T21:42:36Z
|
https://github.com/timkpaine/lantern/issues/201
|
[
"feature",
"backlog"
] |
timkpaine
| 0
|
|
fastapi/sqlmodel
|
sqlalchemy
| 95
|
How to deal with column type needed situation?
|
### First Check
- [X] I added a very descriptive title to this issue.
- [X] I used the GitHub search to find a similar issue and didn't find it.
- [X] I searched the SQLModel documentation, with the integrated search.
- [X] I already searched in Google "How to X in SQLModel" and didn't find any information.
- [X] I already read and followed all the tutorial in the docs and didn't find an answer.
- [X] I already checked if it is not related to SQLModel but to [Pydantic](https://github.com/samuelcolvin/pydantic).
- [X] I already checked if it is not related to SQLModel but to [SQLAlchemy](https://github.com/sqlalchemy/sqlalchemy).
### Commit to Help
- [X] I commit to help with one of those options 👆
### Example Code
```python
from sqlmodel import SQLModel, UniqueConstraint, select
class BookCollection(SQLModel, table=True):
user_nid: int
book_nid: int
UniqueConstraint(
BookCollection.user_nid, # Argument of type "int" cannot be assigned to parameter "columns" of type "str | Column[Any]" in function "__init__"
BookCollection.book_nid, # Argument of type "int" cannot be assigned to parameter "columns" of type "str | Column[Any]" in function "__init__"
name="uidx_book_collection_user_nid_book_nid",
)
# Cannot access member "not_in" for type "int"
select(BookCollection).where(BookCollection.book_nid.not_in([1, 2, 3]))
```
### Description
Some APIs of sqlalchemy may still need a column type. Without that, a type checker will complain.
Currently, I'm using `type: ignore` to skip those.
### Operating System
Linux, macOS
### Operating System Details
_No response_
### SQLModel Version
0.0.4
### Python Version
3.8.6
### Additional Context
_No response_
|
open
|
2021-09-14T08:05:54Z
|
2023-09-03T20:43:36Z
|
https://github.com/fastapi/sqlmodel/issues/95
|
[
"question"
] |
Ma233
| 1
|
Anjok07/ultimatevocalremovergui
|
pytorch
| 757
|
Fail: "[ONNXRuntimeError]
|
Last Error Received:
Process: Ensemble Mode
If this error persists, please contact the developers with the error details.
Raw Error Details:
Fail: "[ONNXRuntimeError] : 1 : FAIL : Non-zero status code returned while running FusedConv node. Name:'Conv_0' Status Message: CUDNN error executing cudnnFindConvolutionForwardAlgorithmEx( s_.handle, s_.x_tensor, s_.x_data, s_.w_desc, s_.w_data, s_.conv_desc, s_.y_tensor, s_.y_data, 1, &algo_count, &perf, algo_search_workspace.get(), max_ws_size)"
Traceback Error: "
File "UVR.py", line 4716, in process_start
File "separate.py", line 287, in seperate
File "separate.py", line 366, in demix_base
File "separate.py", line 386, in run_model
File "separate.py", line 281, in <lambda>
File "onnxruntime\capi\onnxruntime_inference_collection.py", line 192, in run
"
Error Time Stamp [2023-08-22 20:55:57]
Full Application Settings:
vr_model: Choose Model
aggression_setting: 10
window_size: 512
batch_size: Default
crop_size: 256
is_tta: False
is_output_image: False
is_post_process: False
is_high_end_process: False
post_process_threshold: 0.2
vr_voc_inst_secondary_model: No Model Selected
vr_other_secondary_model: No Model Selected
vr_bass_secondary_model: No Model Selected
vr_drums_secondary_model: No Model Selected
vr_is_secondary_model_activate: False
vr_voc_inst_secondary_model_scale: 0.9
vr_other_secondary_model_scale: 0.7
vr_bass_secondary_model_scale: 0.5
vr_drums_secondary_model_scale: 0.5
demucs_model: Choose Model
segment: Default
overlap: 0.25
shifts: 3
chunks_demucs: Auto
margin_demucs: 44100
is_chunk_demucs: False
is_chunk_mdxnet: False
is_primary_stem_only_Demucs: False
is_secondary_stem_only_Demucs: False
is_split_mode: True
is_demucs_combine_stems: True
demucs_voc_inst_secondary_model: No Model Selected
demucs_other_secondary_model: No Model Selected
demucs_bass_secondary_model: No Model Selected
demucs_drums_secondary_model: No Model Selected
demucs_is_secondary_model_activate: False
demucs_voc_inst_secondary_model_scale: 0.9
demucs_other_secondary_model_scale: 0.7
demucs_bass_secondary_model_scale: 0.5
demucs_drums_secondary_model_scale: 0.5
demucs_pre_proc_model: MDX-Net: UVR-MDX-NET-Inst_HQ_3
is_demucs_pre_proc_model_activate: True
is_demucs_pre_proc_model_inst_mix: False
mdx_net_model: Choose Model
chunks: Auto
margin: 44100
compensate: Auto
is_denoise: True
is_invert_spec: True
is_mixer_mode: False
mdx_batch_size: Default
mdx_voc_inst_secondary_model: No Model Selected
mdx_other_secondary_model: No Model Selected
mdx_bass_secondary_model: No Model Selected
mdx_drums_secondary_model: No Model Selected
mdx_is_secondary_model_activate: False
mdx_voc_inst_secondary_model_scale: 0.9
mdx_other_secondary_model_scale: 0.7
mdx_bass_secondary_model_scale: 0.5
mdx_drums_secondary_model_scale: 0.5
is_save_all_outputs_ensemble: True
is_append_ensemble_name: False
chosen_audio_tool: Manual Ensemble
choose_algorithm: Min Spec
time_stretch_rate: 2.0
pitch_rate: 2.0
is_gpu_conversion: True
is_primary_stem_only: False
is_secondary_stem_only: True
is_testing_audio: False
is_add_model_name: True
is_accept_any_input: True
is_task_complete: False
is_normalization: True
is_create_model_folder: False
mp3_bit_set: 320k
save_format: WAV
wav_type_set: PCM_24
help_hints_var: True
model_sample_mode: False
model_sample_mode_duration: 30
demucs_stems: All Stems
I don't know wtf, and I'm not professional in this.
|
open
|
2023-08-22T13:58:07Z
|
2023-08-22T19:10:34Z
|
https://github.com/Anjok07/ultimatevocalremovergui/issues/757
|
[] |
dimadt
| 3
|
tartiflette/tartiflette
|
graphql
| 256
|
.cook() is not idempotent
|
* [x] **Explain with a simple sentence the expected behavior**: it should be possible to call `.cook()` multiple times without it failing with hard-to-debug error tracebacks.
* [x] **Tartiflette version:** 0.11.2
* [x] **Python version:** 3.7.2
* [x] **Executed in docker:** No
* [x] **Is it a regression from a previous versions?** No (I think the root cause has always been there?)
* [x] **Stack trace**: I pushed a failing test in #255
```python
_______________________________________ ERROR at setup of test_error_handling _______________________________________
ttftt = <tartiflette_starlette.app.TartifletteApp object at 0x105b25668>
@pytest.fixture(name="client")
def fixture_client(ttftt: TartifletteApp) -> TestClient:
> with TestClient(ttftt) as client:
tests/conftest.py:28:
_ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _
/Users/Florimond/Developer/tartiflette-projects/tartiflette-starlette/venv/lib/python3.7/site-packages/starlette/testclient.py:449: in __enter__
loop.run_until_complete(self.wait_startup())
/Users/Florimond/.pyenv/versions/3.7.2/lib/python3.7/asyncio/base_events.py:584: in run_until_complete
return future.result()
/Users/Florimond/Developer/tartiflette-projects/tartiflette-starlette/venv/lib/python3.7/site-packages/starlette/testclient.py:467: in wait_startup
self.task.result()
/Users/Florimond/Developer/tartiflette-projects/tartiflette-starlette/venv/lib/python3.7/site-packages/starlette/testclient.py:459: in lifespan
await self.app(scope, self.receive_queue.get, self.send_queue.put)
tartiflette_starlette/app.py:55: in __call__
await self.lifespan(scope, receive, send)
/Users/Florimond/Developer/tartiflette-projects/tartiflette-starlette/venv/lib/python3.7/site-packages/starlette/routing.py:472: in __call__
await self.startup()
/Users/Florimond/Developer/tartiflette-projects/tartiflette-starlette/venv/lib/python3.7/site-packages/starlette/routing.py:458: in startup
await handler()
tartiflette_starlette/app.py:51: in on_startup
await self.engine.cook()
/Users/Florimond/Developer/tartiflette-projects/tartiflette-starlette/venv/lib/python3.7/site-packages/tartiflette/engine.py:157: in cook
self._modules, modules_sdl = await _import_modules(modules, schema_name)
/Users/Florimond/Developer/tartiflette-projects/tartiflette-starlette/venv/lib/python3.7/site-packages/tartiflette/engine.py:76: in _import_modules
module = import_module(module["name"])
_ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _
name = <module 'tests.resolvers' from '/Users/florimond/Developer/tartiflette-projects/tartiflette-starlette/tests/resolvers.py'>
package = None
def import_module(name, package=None):
"""Import a module.
The 'package' argument is required when performing a relative import. It
specifies the package to use as the anchor point from which to resolve the
relative import to an absolute import.
"""
level = 0
> if name.startswith('.'):
E AttributeError: module 'tests.resolvers' has no attribute 'startswith'
/Users/Florimond/.pyenv/versions/3.7.2/lib/python3.7/importlib/__init__.py:118: AttributeError
```
**More details**
While playing with https://github.com/tartiflette/tartiflette-starlette/issues/32 I ran into issues when `.cook()` was being called multiple times. (I need to to this because we can't create an `Engine` multiple times in the same session, and so one single `Engine` is reused across tests.)
Ultimately, here are the reasons why I think this doesn't work:
- `.cook()` tries to import modules, although `self._modules` might already contain the imported `module` objects from a previous call. Overall I think this (and potentially other issues) can be fixed by adding a simple `._cooked` flag and skipping `.cook()` if it's `True`.
- `.cook()` always registers the SDL, even though it may have already been added to the `SchemaRegistry` before, which fails on second time because the same SDL is used, resulting in conflicting types. (I suppose this is why we use a `clean_registry` in the engine unit tests.) I don't really have an idea on how to fix this. Should I just clean the registry like we do in the tests before calling `.cook()` in `tartiflette-starlette`?
|
closed
|
2019-07-02T07:49:57Z
|
2019-07-02T22:30:07Z
|
https://github.com/tartiflette/tartiflette/issues/256
|
[
"enhancement"
] |
florimondmanca
| 8
|
vanna-ai/vanna
|
data-visualization
| 228
|
Ask Snowflake - Couldn't run sql: 'NoneType' object has no attribute 'fetchall'
|
I am running vn.ask(question="some natual language questions") to query Snowflake table, and it returns the following error.
None
WARNING:snowflake.connector.cursor:execute: no query is given to execute
Couldn't run sql: 'NoneType' object has no attribute 'fetchall'
|
closed
|
2024-02-02T10:29:33Z
|
2024-03-02T04:13:22Z
|
https://github.com/vanna-ai/vanna/issues/228
|
[] |
boyuanqian
| 0
|
hankcs/HanLP
|
nlp
| 1,162
|
关于简繁转换的问题
|
<!--
注意事项和版本号必填,否则不回复。若希望尽快得到回复,请按模板认真填写,谢谢合作。
-->
## 注意事项
请确认下列注意事项:
* 我已仔细阅读下列文档,都没有找到答案:
- [首页文档](https://github.com/hankcs/HanLP)
- [wiki](https://github.com/hankcs/HanLP/wiki)
- [常见问题](https://github.com/hankcs/HanLP/wiki/FAQ)
* 我已经通过[Google](https://www.google.com/#newwindow=1&q=HanLP)和[issue区检索功能](https://github.com/hankcs/HanLP/issues)搜索了我的问题,也没有找到答案。
* 我明白开源社区是出于兴趣爱好聚集起来的自由社区,不承担任何责任或义务。我会礼貌发言,向每一个帮助我的人表示感谢。
* [x] 我在此括号内输入x打钩,代表上述事项确认完毕。
## 版本号
1.6.5
## 我的问题
System.out.println(HanLP.convertToSimplifiedChinese("「以後等妳當上皇后,就能買士多啤梨慶祝了」"));
输出 : “以后等你当上皇后,就能买草莓庆祝了”
System.out.println(HanLP.convertToTraditionalChinese("“以后等你当上皇后,就能买草莓庆祝了”"));
输出 : “以後等你當上皇后,就能買草莓慶祝了”
期望能输出 “以後等妳當上皇后,就能買士多啤梨慶祝了”,是有什么地方需要配置吗?
|
closed
|
2019-04-26T07:19:26Z
|
2019-04-26T07:38:17Z
|
https://github.com/hankcs/HanLP/issues/1162
|
[] |
humiao8sz
| 1
|
python-visualization/folium
|
data-visualization
| 1,286
|
How to open the popup automatically when a search is found for a GeoJson layer?
|
#### Please add a code sample or a nbviewer link, copy-pastable if possible
```python
# Your code here
import folium
import branca
import geopandas
from folium.plugins import Search
print(folium.__version__)
states = geopandas.read_file(
'https://rawcdn.githack.com/PublicaMundi/MappingAPI/master/data/geojson/us-states.json',
driver='GeoJSON'
)
min, max = states['density'].quantile([0.05,0.95]).apply(lambda x: round(x, 2))
colormap = branca.colormap.LinearColormap(
colors=['#f2f0f7','#cbc9e2','#9e9ac8','#756bb1','#54278f'],
index=states['density'].quantile([0.2,0.4,0.6,0.8]),
vmin=min,
vmax=max
)
colormap.caption="Population Density in the United States"
m = folium.Map(location=[38,-97], zoom_start=4)
style_function = lambda x: {
'fillColor': colormap(x['properties']['density']),
'color': 'black',
'weight':2,
'fillOpacity':0.5
}
stategeo = folium.GeoJson(
states,
name='US States',
style_function=style_function,
popup=folium.GeoJsonPopup(
fields=['name', 'density'],
aliases=['State', 'Density'],
localize=True
)
).add_to(m)
statesearch = Search(
layer=stategeo,
geom_type='Polygon',
placeholder='Search for a US State',
collapsed=False,
search_label='name'
).add_to(m)
folium.LayerControl().add_to(m)
colormap.add_to(m)
m
```
#### Problem description
I was trying to make the popup open automatically when a search is found in a GeoJson layer. My code is modified from an example code "plugin-Search.ipynb". I am using the "master" version of folium on the github. Right now, my code cannot open the popup when a search is found. I checked the source code "search.py" in "plugins" folder. The code is:
{{this.layer.get_name()}}searchControl.on('search:locationfound', function(e) {
{{this.layer.get_name()}}.setStyle(function(feature){
return feature.properties.style
})
{% if this.options %}
e.layer.setStyle({{ this.options|tojson }});
{% endif %}
if(e.layer._popup)
e.layer.openPopup();
})
I think the code should open popup if location is found but it did not.
I manually added console.log(e.layer._popup) in front of if(e.layer._popup) in the html file. When I searched one state on the web page, the console outputs "undefined" for e.layer._popup. It's confusing to me.
#### Expected Output
I expect to open the popup automatically when a search is found like this leaflet search example https://labs.easyblog.it/maps/leaflet-search/examples/geojson-layer.html.
#### Output of ``folium.__version__``
0+unknown
|
open
|
2020-04-10T04:18:51Z
|
2022-11-28T16:56:55Z
|
https://github.com/python-visualization/folium/issues/1286
|
[
"bug"
] |
RenchengDong
| 1
|
onnx/onnx
|
pytorch
| 6,013
|
AttributeError while installing ONNX
|
# Bug Report
### Is the issue related to model conversion?
<!-- If the ONNX checker reports issues with this model then this is most probably related to the converter used to convert the original framework model to ONNX. Please create this bug in the appropriate converter's GitHub repo (pytorch, tensorflow-onnx, sklearn-onnx, keras-onnx, onnxmltools) to get the best help. -->
No
### Describe the bug
<!-- Please describe the bug clearly and concisely -->
when I install ONNX from source in Ubuntu22.04, I follow the instructions in [install ONNX in Linux](https://github.com/onnx/onnx?tab=readme-ov-file#linux). However it failed.
### System information
<!--
- OS Platform and Distribution (*e.g. Linux Ubuntu 20.04*):
- ONNX version (*e.g. 1.13*):
- Python version:
- GCC/Compiler version (if compiling from source):
- CMake version:
- Protobuf version:
- Visual Studio version (if applicable):-->
- OS: Ubuntu22.04
- ONNX: latest
- python: 3.10.12
- GCC: 11.4.0
- Cmake: 3.22.1
- Protobuf: 3.21.12
### Reproduction instructions
<!--
- Describe the code to reproduce the behavior.
```
import onnx
model = onnx.load('model.onnx')
...
```
- Attach the ONNX model to the issue (where applicable)-->
just follow the [install ONNX in Linux](https://github.com/onnx/onnx?tab=readme-ov-file#linux)
### Error logs
```
➜ onnx git:(main) ✗ sudo pip install .
Processing /home/cwwu/Downloads/onnx
Installing build dependencies ... done
Getting requirements to build wheel ... done
Installing backend dependencies ... done
Preparing metadata (pyproject.toml) ... done
Building wheels for collected packages: UNKNOWN
Building wheel for UNKNOWN (pyproject.toml) ... error
error: subprocess-exited-with-error
× Building wheel for UNKNOWN (pyproject.toml) did not run successfully.
│ exit code: 1
╰─> [105 lines of output]
running bdist_wheel
running build
running build_ext
running cmake_build
-- ONNX_PROTOC_EXECUTABLE: /usr/bin/protoc
-- Protobuf_VERSION: 3.21.12
Generated: /home/cwwu/Downloads/onnx/.setuptools-cmake-build/onnx/onnx-ml.proto
Generated: /home/cwwu/Downloads/onnx/.setuptools-cmake-build/onnx/onnx-operators-ml.proto
Generated: /home/cwwu/Downloads/onnx/.setuptools-cmake-build/onnx/onnx-data.proto
-- Could NOT find pybind11 (missing: pybind11_DIR)
-- pybind11 v2.10.4
--
-- ******** Summary ********
-- CMake version : 3.22.1
-- CMake command : /usr/bin/cmake
-- System : Linux
-- C++ compiler : /usr/bin/c++
-- C++ compiler version : 11.4.0
-- CXX flags : -Wnon-virtual-dtor
-- Build type : Release
-- Compile definitions : __STDC_FORMAT_MACROS
-- CMAKE_PREFIX_PATH :
-- CMAKE_INSTALL_PREFIX : /usr/local
-- CMAKE_MODULE_PATH :
--
-- ONNX version : 1.17.0
-- ONNX NAMESPACE : onnx
-- ONNX_USE_LITE_PROTO : OFF
-- USE_PROTOBUF_SHARED_LIBS : OFF
-- Protobuf_USE_STATIC_LIBS : ON
-- ONNX_DISABLE_EXCEPTIONS : OFF
-- ONNX_DISABLE_STATIC_REGISTRATION : OFF
-- ONNX_WERROR : OFF
-- ONNX_BUILD_TESTS : OFF
-- ONNX_BUILD_BENCHMARKS : OFF
-- ONNX_BUILD_SHARED_LIBS :
-- BUILD_SHARED_LIBS :
--
-- Protobuf compiler : /usr/bin/protoc
-- Protobuf includes : /usr/include
-- Protobuf libraries : /usr/lib/x86_64-linux-gnu/libprotobuf.a
-- BUILD_ONNX_PYTHON : ON
-- Python version :
-- Python executable : /usr/bin/python3
-- Python includes : /usr/include/python3.10
-- Configuring done
-- Generating done
-- Build files have been written to: /home/cwwu/Downloads/onnx/.setuptools-cmake-build
[ 2%] Built target gen_onnx_proto
[ 8%] Built target gen_onnx_operators_proto
[ 8%] Built target gen_onnx_data_proto
Consolidate compiler generated dependencies of target onnx_proto
[ 22%] Built target onnx_proto
Consolidate compiler generated dependencies of target onnx
[ 97%] Built target onnx
Consolidate compiler generated dependencies of target onnx_cpp2py_export
[100%] Built target onnx_cpp2py_export
Traceback (most recent call last):
File "/usr/lib/python3/dist-packages/pip/_vendor/pep517/in_process/_in_process.py", line 363, in <module>
main()
File "/usr/lib/python3/dist-packages/pip/_vendor/pep517/in_process/_in_process.py", line 345, in main
json_out['return_val'] = hook(**hook_input['kwargs'])
File "/usr/lib/python3/dist-packages/pip/_vendor/pep517/in_process/_in_process.py", line 261, in build_wheel
return _build_backend().build_wheel(wheel_directory, config_settings,
File "/usr/lib/python3/dist-packages/setuptools/build_meta.py", line 230, in build_wheel
return self._build_with_temp_dir(['bdist_wheel'], '.whl',
File "/usr/lib/python3/dist-packages/setuptools/build_meta.py", line 215, in _build_with_temp_dir
self.run_setup()
File "/usr/lib/python3/dist-packages/setuptools/build_meta.py", line 158, in run_setup
exec(compile(code, __file__, 'exec'), locals())
File "setup.py", line 309, in <module>
setuptools.setup(
File "/usr/lib/python3/dist-packages/setuptools/__init__.py", line 153, in setup
return distutils.core.setup(**attrs)
File "/usr/lib/python3/dist-packages/setuptools/_distutils/core.py", line 148, in setup
return run_commands(dist)
File "/usr/lib/python3/dist-packages/setuptools/_distutils/core.py", line 163, in run_commands
dist.run_commands()
File "/usr/lib/python3/dist-packages/setuptools/_distutils/dist.py", line 967, in run_commands
self.run_command(cmd)
File "/usr/lib/python3/dist-packages/setuptools/_distutils/dist.py", line 986, in run_command
cmd_obj.run()
File "/usr/lib/python3/dist-packages/wheel/bdist_wheel.py", line 299, in run
self.run_command('build')
File "/usr/lib/python3/dist-packages/setuptools/_distutils/cmd.py", line 313, in run_command
self.distribution.run_command(command)
File "/usr/lib/python3/dist-packages/setuptools/_distutils/dist.py", line 986, in run_command
cmd_obj.run()
File "/usr/lib/python3/dist-packages/setuptools/_distutils/command/build.py", line 135, in run
self.run_command(cmd_name)
File "/usr/lib/python3/dist-packages/setuptools/_distutils/cmd.py", line 313, in run_command
self.distribution.run_command(command)
File "/usr/lib/python3/dist-packages/setuptools/_distutils/dist.py", line 986, in run_command
cmd_obj.run()
File "setup.py", line 247, in run
return super().run()
File "/usr/lib/python3/dist-packages/setuptools/command/build_ext.py", line 79, in run
_build_ext.run(self)
File "/usr/lib/python3/dist-packages/setuptools/_distutils/command/build_ext.py", line 339, in run
self.build_extensions()
File "setup.py", line 276, in build_extensions
if self.editable_mode:
File "/usr/lib/python3/dist-packages/setuptools/_distutils/cmd.py", line 103, in __getattr__
raise AttributeError(attr)
AttributeError: editable_mode
[end of output]
note: This error originates from a subprocess, and is likely not a problem with pip.
ERROR: Failed building wheel for UNKNOWN
Failed to build UNKNOWN
ERROR: Could not build wheels for UNKNOWN, which is required to install pyproject.toml-based projects
```
|
closed
|
2024-03-11T14:02:25Z
|
2024-07-31T16:01:36Z
|
https://github.com/onnx/onnx/issues/6013
|
[
"question",
"topic: build"
] |
Tom-Teamo
| 13
|
svc-develop-team/so-vits-svc
|
pytorch
| 126
|
[Help]: 每次训练都在 Epoch: 2 [42%], step: 800 的位置报错退出
|
### 请勾选下方的确认框。
- [x] 我已仔细阅读[README.md](https://github.com/svc-develop-team/so-vits-svc/blob/4.0/README_zh_CN.md)和[wiki中的Quick solution](https://github.com/svc-develop-team/so-vits-svc/wiki/Quick-solution)。
- [X] 我已通过各种搜索引擎排查问题,我要提出的问题并不常见。
- [X] 我未在使用由第三方用户提供的一键包/环境包。
### 系统平台版本号
Windows 10 家庭版
### GPU 型号
NVIDIA GeForce RTX 2060
### Python版本
python3.9.7
### PyTorch版本
2.0.0+cu118
### sovits分支
4.0-v2
### 数据集来源(用于判断数据集质量)
动画原声采集
### 出现问题的环节或执行的命令
训练
### 问题描述
每次训练都在 INFO:44k:Train Epoch: 2 [42%], step: 800, 的位置报错退出
即使把logs/44k文件夹清空重新训练,也是一样在这个位置报错退出
由于显卡是6G显存,config.json中我修改了batch_size,从6改为2 ,"batch_size": 2
### 日志
```python
INFO:44k:Loaded checkpoint './logs\44k\D_0.pth' (iteration 1)
E:\Python\lib\site-packages\torch\functional.py:641: UserWarning: stft with return_complex=False is deprecated. In a future pytorch release, stft will return complex tensors for all inputs, and return_complex=False will raise an error.
Note: you can still call torch.view_as_real on the complex output to recover the old return format. (Triggered internally at C:\actions-runner\_work\pytorch\pytorch\builder\windows\pytorch\aten\src\ATen\native\SpectralOps.cpp:867.)
return _VF.stft(input, n_fft, hop_length, win_length, window, # type: ignore[attr-defined]
INFO:torch.nn.parallel.distributed:Reducer buckets have been rebuilt in this iteration.
E:\Python\lib\site-packages\torch\autograd\__init__.py:200: UserWarning: Grad strides do not match bucket view strides. This may indicate grad was not created according to the gradient layout contract, or that the param's strides changed since DDP was constructed. This is not an error, but may impair performance.
grad.sizes() = [32, 1, 4], strides() = [4, 1, 1]
bucket_view.sizes() = [32, 1, 4], strides() = [4, 4, 1] (Triggered internally at C:\actions-runner\_work\pytorch\pytorch\builder\windows\pytorch\torch\csrc\distributed\c10d\reducer.cpp:337.)
Variable._execution_engine.run_backward( # Calls into the C++ engine to run the backward pass
INFO:44k:Train Epoch: 1 [0%]
INFO:44k:Losses: [1.8350424766540527, 3.087881326675415, 15.519474029541016, 43.46516799926758, 2.739884614944458], step: 0, lr: 0.0001
INFO:44k:Saving model and optimizer state at iteration 1 to ./logs\44k\G_0.pth
INFO:44k:Saving model and optimizer state at iteration 1 to ./logs\44k\D_0.pth
INFO:torch.nn.parallel.distributed:Reducer buckets have been rebuilt in this iteration.
INFO:44k:Train Epoch: 1 [35%]
INFO:44k:Losses: [3.2042624950408936, 1.6023280620574951, 4.85037899017334, 24.364368438720703, 1.8815621137619019], step: 200, lr: 0.0001
INFO:44k:Train Epoch: 1 [71%]
INFO:44k:Losses: [2.024649143218994, 2.5297534465789795, 12.023140907287598, 29.055946350097656, 1.7351597547531128], step: 400, lr: 0.0001
INFO:44k:====> Epoch: 1, cost 366.75 s
INFO:44k:Train Epoch: 2 [6%]
INFO:44k:Losses: [1.9109034538269043, 3.0717015266418457, 15.881654739379883, 31.35890769958496, 2.1832027435302734], step: 600, lr: 9.99875e-05
INFO:44k:Train Epoch: 2 [42%]
INFO:44k:Losses: [2.1936745643615723, 2.2962450981140137, 8.9740629196167, 23.723608016967773, 1.4403268098831177], step: 800, lr: 9.99875e-05
Traceback (most recent call last):
File "E:\1\2\so-vits-svc\train.py", line 315, in <module>
main()
File "E:\1\2\so-vits-svc\train.py", line 53, in main
mp.spawn(run, nprocs=n_gpus, args=(n_gpus, hps,))
File "E:\Python\lib\site-packages\torch\multiprocessing\spawn.py", line 239, in spawn
return start_processes(fn, args, nprocs, join, daemon, start_method='spawn')
File "E:\Python\lib\site-packages\torch\multiprocessing\spawn.py", line 197, in start_processes
while not context.join():
File "E:\Python\lib\site-packages\torch\multiprocessing\spawn.py", line 160, in join
raise ProcessRaisedException(msg, error_index, failed_process.pid)
torch.multiprocessing.spawn.ProcessRaisedException:
-- Process 0 terminated with the following error:
Traceback (most recent call last):
File "E:\Python\lib\site-packages\torch\utils\data\dataloader.py", line 1133, in _try_get_data
data = self._data_queue.get(timeout=timeout)
File "E:\Python\lib\multiprocessing\queues.py", line 114, in get
raise Empty
_queue.Empty
The above exception was the direct cause of the following exception:
Traceback (most recent call last):
File "E:\Python\lib\site-packages\torch\multiprocessing\spawn.py", line 69, in _wrap
fn(i, *args)
File "E:\1\2\so-vits-svc\train.py", line 124, in run
train_and_evaluate(rank, epoch, hps, [net_g, net_d], [optim_g, optim_d], [scheduler_g, scheduler_d], scaler,
File "E:\1\2\so-vits-svc\train.py", line 245, in train_and_evaluate
evaluate(hps, net_g, eval_loader, writer_eval)
File "E:\1\2\so-vits-svc\train.py", line 269, in evaluate
for batch_idx, items in enumerate(eval_loader):
File "E:\Python\lib\site-packages\torch\utils\data\dataloader.py", line 634, in __next__
data = self._next_data()
File "E:\Python\lib\site-packages\torch\utils\data\dataloader.py", line 1329, in _next_data
idx, data = self._get_data()
File "E:\Python\lib\site-packages\torch\utils\data\dataloader.py", line 1295, in _get_data
success, data = self._try_get_data()
File "E:\Python\lib\site-packages\torch\utils\data\dataloader.py", line 1146, in _try_get_data
raise RuntimeError('DataLoader worker (pid(s) {}) exited unexpectedly'.format(pids_str)) from e
RuntimeError: DataLoader worker (pid(s) 18212) exited unexpectedly
```
### 截图`so-vits-svc`、`logs/44k`文件夹并粘贴到此处

### 补充说明
_No response_
|
closed
|
2023-04-05T18:59:38Z
|
2023-04-13T04:03:13Z
|
https://github.com/svc-develop-team/so-vits-svc/issues/126
|
[
"help wanted"
] |
AGuanDao
| 5
|
jina-ai/serve
|
machine-learning
| 5,744
|
Torch error in Jcloud http flow
|
**Describe the bug**
<!-- A clear and concise description of what the bug is. -->
Posting to the below flow in jcloud http protocol will throw a torch module not found error. Grpc is fine.
```
from jina import DocumentArray, Executor, requests
import torch
class dummy_torch(Executor):
@requests
def foo(self, docs: DocumentArray, **kwargs):
for d in docs:
d.embedding = torch.rand(1000)
```
YAML
```
jtype: Flow
with:
prefetch: 5
gateway:
port:
- 51000
- 52000
protocol:
- grpc
- http
executors:
- name: dummyExecutor
uses: jinaai+docker://auth0-unified-40be9bf07eece29a/dummy_torch:latest
env:
JINA_LOG_LEVEL: DEBUG
```
LOCAL:
```
from jina import DocumentArray, Client
client = Client(host='jcloud-endpoint)
res = client.post(on='/', inputs=DocumentArray.empty(5), show_progress=True)
res.summary()
```
LOCAL error trace:
```
Traceback (most recent call last):
File "toy.py", line 5, in <module>
res = client.post(on='/', inputs=DocumentArray.empty(5), show_progress=True)
File "/Users/ziniuyu/Documents/github/jina/jina/clients/mixin.py", line 281, in post
return run_async(
File "/Users/ziniuyu/Documents/github/jina/jina/helper.py", line 1334, in run_async
return asyncio.run(func(*args, **kwargs))
File "/opt/anaconda3/envs/py3813/lib/python3.8/asyncio/runners.py", line 44, in run
return loop.run_until_complete(main)
File "/opt/anaconda3/envs/py3813/lib/python3.8/asyncio/base_events.py", line 616, in run_until_complete
return future.result()
File "/Users/ziniuyu/Documents/github/jina/jina/clients/mixin.py", line 271, in _get_results
async for resp in c._get_results(*args, **kwargs):
File "/Users/ziniuyu/Documents/github/jina/jina/clients/base/http.py", line 165, in _get_results
r_str = await response.json()
File "/opt/anaconda3/envs/py3813/lib/python3.8/site-packages/aiohttp/client_reqrep.py", line 1104, in json
raise ContentTypeError(
aiohttp.client_exceptions.ContentTypeError: 0, message='Attempt to decode JSON with unexpected mimetype: text/plain; charset=utf-8', url=URL('jcloud-endpoint:443/post')
```
Gateway error trace:
```
ERROR: Exception in ASGI application
Traceback (most recent call last):
File "/usr/local/lib/python3.8/site-packages/uvicorn/protocols/http/httptools_impl.py", line 419, in run_asgi
result = await app( # type: ignore[func-returns-value]
File "/usr/local/lib/python3.8/site-packages/uvicorn/middleware/proxy_headers.py", line 78, in __call__
return await self.app(scope, receive, send)
File "/usr/local/lib/python3.8/site-packages/fastapi/applications.py", line 271, in __call__
await super().__call__(scope, receive, send)
File "/usr/local/lib/python3.8/site-packages/starlette/applications.py", line 118, in __call__
await self.middleware_stack(scope, receive, send)
File "/usr/local/lib/python3.8/site-packages/starlette/middleware/errors.py", line 184, in __call__
raise exc
File "/usr/local/lib/python3.8/site-packages/starlette/middleware/errors.py", line 162, in __call__
await self.app(scope, receive, _send)
File "/usr/local/lib/python3.8/site-packages/starlette/middleware/exceptions.py", line 79, in __call__
raise exc
File "/usr/local/lib/python3.8/site-packages/starlette/middleware/exceptions.py", line 68, in __call__
await self.app(scope, receive, sender)
File "/usr/local/lib/python3.8/site-packages/fastapi/middleware/asyncexitstack.py", line 21, in __call__
raise e
File "/usr/local/lib/python3.8/site-packages/fastapi/middleware/asyncexitstack.py", line 18, in __call__
await self.app(scope, receive, send)
File "/usr/local/lib/python3.8/site-packages/starlette/routing.py", line 706, in __call__
await route.handle(scope, receive, send)
File "/usr/local/lib/python3.8/site-packages/starlette/routing.py", line 276, in handle
await self.app(scope, receive, send)
File "/usr/local/lib/python3.8/site-packages/starlette/routing.py", line 66, in app
response = await func(request)
File "/usr/local/lib/python3.8/site-packages/fastapi/routing.py", line 237, in app
raw_response = await run_endpoint_function(
File "/usr/local/lib/python3.8/site-packages/fastapi/routing.py", line 163, in run_endpoint_function
return await dependant.call(**values)
File "/usr/local/lib/python3.8/site-packages/jina/serve/runtimes/gateway/http/app.py", line 191, in post
result = await _get_singleton_result(
File "/usr/local/lib/python3.8/site-packages/jina/serve/runtimes/gateway/http/app.py", line 382, in _get_singleton_result
result_dict = result.to_dict()
File "/usr/local/lib/python3.8/site-packages/jina/types/request/data.py", line 260, in to_dict
da = self.docs
File "/usr/local/lib/python3.8/site-packages/jina/types/request/data.py", line 276, in docs
return self.data.docs
File "/usr/local/lib/python3.8/site-packages/jina/types/request/data.py", line 47, in docs
self._loaded_doc_array = self.document_array_cls.from_protobuf(
File "/usr/local/lib/python3.8/site-packages/docarray/array/mixins/io/binary.py", line 361, in from_protobuf
return cls(Document.from_protobuf(od) for od in pb_msg.docs)
File "/usr/local/lib/python3.8/site-packages/docarray/array/mixins/io/from_gen.py", line 23, in __init__
super().__init__(*args, **kwargs)
File "/usr/local/lib/python3.8/site-packages/docarray/array/base.py", line 12, in __init__
self._init_storage(*args, **kwargs)
File "/usr/local/lib/python3.8/site-packages/docarray/array/storage/memory/backend.py", line 25, in wrapper
return func(self, *args, **kwargs)
File "/usr/local/lib/python3.8/site-packages/docarray/array/storage/memory/backend.py", line 83, in _init_storage
self.extend(_docs)
File "/usr/local/lib/python3.8/site-packages/docarray/array/storage/base/seqlike.py", line 81, in extend
self._extend(values, **kwargs)
File "/usr/local/lib/python3.8/site-packages/docarray/array/storage/memory/seqlike.py", line 60, in _extend
values = list(values) # consume the iterator only once
File "/usr/local/lib/python3.8/site-packages/docarray/array/mixins/io/binary.py", line 361, in <genexpr>
return cls(Document.from_protobuf(od) for od in pb_msg.docs)
File "/usr/local/lib/python3.8/site-packages/docarray/document/mixins/protobuf.py", line 13, in from_protobuf
return parse_proto(pb_msg)
File "/usr/local/lib/python3.8/site-packages/docarray/proto/io/__init__.py", line 24, in parse_proto
fields[f_name] = read_ndarray(value)
File "/usr/local/lib/python3.8/site-packages/docarray/proto/io/ndarray.py", line 44, in read_ndarray
return _to_framework_array(x, framework)
File "/usr/local/lib/python3.8/site-packages/docarray/proto/io/ndarray.py", line 147, in _to_framework_array
from torch import from_numpy
ModuleNotFoundError: No module named 'torch'
```
**Describe how you solve it**
<!-- copy past your code/pull request link -->
No error with numpy
Possible reason: the default http gateway image does not have torch installed
---
<!-- Optional, but really help us locate the problem faster -->
**Environment**
<!-- Run `jina --version-full` and copy paste the output here -->
**Screenshots**
<!-- If applicable, add screenshots to help explain your problem. -->
|
closed
|
2023-03-07T12:01:10Z
|
2023-03-13T21:55:46Z
|
https://github.com/jina-ai/serve/issues/5744
|
[] |
ZiniuYu
| 0
|
nolar/kopf
|
asyncio
| 966
|
Replay resources doesn't work
|
### Keywords
_No response_
### Problem
Hello, my use case manages two CRDs (DNS instance and DNS records). My operator allows removing the DNS instance leaving the associated DNS records present just in case it obeys to a temporary need.
After recreating the DNS instance I planned to simply run "kubectl get dnsr -o yaml | kubectl replace -f -" to reprocess them but I got a strange behaviour. The resources are created but the code associated to the operator is not executed.
After digging into the documentation I realized that "kopf.zalando.org/last-handled-configuration" annotation explains this behaviour.
My question then, is there any better approach than just asking to remove the annotations with something like this?
```
kubectl get dnsr -o yaml | \
yq 'del(.items[].metadata.annotations."kopf.zalando.org/last-handled-configuration")' - | \
kubectl replace -f -
```
Thanks for your help.
|
closed
|
2022-10-30T10:32:38Z
|
2022-11-07T19:14:30Z
|
https://github.com/nolar/kopf/issues/966
|
[
"question"
] |
davidp1404
| 2
|
google-research/bert
|
tensorflow
| 1,148
|
Can RoBERTa be fine-tuned on unlabeled data?
|
Im new working with language models, need some help
Can i pre train RoBERTa with data set having just one column i-e sentences and no labels or anything?
|
open
|
2020-09-16T06:39:13Z
|
2020-09-16T06:39:13Z
|
https://github.com/google-research/bert/issues/1148
|
[] |
HamzaYounis
| 0
|
aeon-toolkit/aeon
|
scikit-learn
| 1,931
|
[ENH] Improvements to write_to_tsfile
|
### Describe the feature or idea you want to propose
just writing some data to file and will note anything here that could be better
### Describe your proposed solution
1. More informative errors
2. Data precision option: it currently creates big files
3. Doesnt work with univariate data in 2D format
|
closed
|
2024-08-08T10:07:40Z
|
2024-11-23T08:46:43Z
|
https://github.com/aeon-toolkit/aeon/issues/1931
|
[
"enhancement",
"datasets"
] |
TonyBagnall
| 0
|
cobrateam/splinter
|
automation
| 570
|
Unexpected behavior from setting profiles and extensions.
|
Python = 2.7.10 (mac OS X Sierra)
Tried:
browser = Browser('firefox', extensions=['/full/path/to/ext.xpi', '/full/path/to/ext2.xpi'])
browser = Browser('firefox', extensions=['./ext.xpi', './ext2.xpi'])
With and without escape characters because it's in 'Application Support'
browser = Browser('firefox', profile=['/full/path/to/thingy.default, '/full/path/to/thing2.second'])
browser = Browser('firefox', profile=['./thingy.default, './thing2.second'])
Again With and without escape characters because it's in 'Application Support'
Even combined both extension and profile. None of it seems to work, always similar errors "[Errno 2] No such file or directory: ".
|
closed
|
2017-11-01T01:52:34Z
|
2021-07-22T21:59:15Z
|
https://github.com/cobrateam/splinter/issues/570
|
[
"NeedsInvestigation"
] |
orange-tsai
| 2
|
noirbizarre/flask-restplus
|
flask
| 466
|
App runs locally but returns 500 error on Heroku
|
There doesn't seem to be any documentation on deploying to Heroku with flask-restplus. I've just deployed an app and am getting the following: `Error: INTERNAL SERVER ERROR`.
My Procfile is set to `web: gunicorn app:app` and my app is set as `api = Api(app)`, `app.wsgi_app = ProxyFix(app.wsgi_app)`, and `app = Flask(__name__)`, respectively. Anyone have any suggestions?
|
closed
|
2018-06-05T15:44:07Z
|
2024-03-20T17:15:35Z
|
https://github.com/noirbizarre/flask-restplus/issues/466
|
[] |
rah-ool
| 39
|
Esri/arcgis-python-api
|
jupyter
| 1,815
|
License.all() returns incorrect results for orgs with >10,000 users
|
In an ArcGIS Online organization with >10,000 users, the License.all() method returns only the first 10,000 users that have a particular license. There does not seem to be a way to page results or otherwise access the users above the first 10,000.
You can reproduce this issue in any org with more than 10,000 users, for something is licensed to more than 10,000 users, for which you attempt to use all() to retrieve the full list of usernames to which it is licensed.
If the 10,000 limitation cannot be addressed, then it would be helpful to have all() return an error message that the list of usernames exceeds the limit and is incomplete, which a script could catch.
In the meantime, any suggestions for a workaround that would help construct the complete list of users that have a license? Is getting a list of all users, and checking them one-by-one for that license, the best way to do it?
|
open
|
2024-04-28T14:56:37Z
|
2024-06-24T18:21:50Z
|
https://github.com/Esri/arcgis-python-api/issues/1815
|
[
"enhancement"
] |
knoopum
| 0
|
waditu/tushare
|
pandas
| 1,617
|
数据获取工具获取上市公司管理层信息报错
|
环境:Chrome浏览器
功能点击:Tushare数据获取工具,获取上市公司管理层 时,报错,错误没有完全显示
缺陷提交人:494037935@qq.com
|
open
|
2022-01-01T12:25:39Z
|
2022-01-01T12:26:02Z
|
https://github.com/waditu/tushare/issues/1617
|
[] |
znufe2010
| 0
|
microsoft/JARVIS
|
pytorch
| 45
|
Would it support Azure OpenAI?
|
Hi, I only saw OpenAI API key in config.yml. Does it support Azure OpenAI? Or is there any plans to support Azure OpenAI?
|
closed
|
2023-04-05T14:51:30Z
|
2023-04-10T05:22:04Z
|
https://github.com/microsoft/JARVIS/issues/45
|
[] |
Ko-Ko-Kirk
| 2
|
pennersr/django-allauth
|
django
| 3,679
|
Assertion Error for Existing Users After Registering New User with Social Account
|
Hello,
I've encountered an issue where, after registering a new user through a social account integration, all existing users are unable to log in and are met with an AssertionError. This problem persists across all stored users and makes it impossible for them to log in after a new user registers via a social account.
Steps to Reproduce:
1. Register a new user using a social account (e.g., Google, Facebook).
2. Attempt to log in with an existing user account.
3. An AssertionError is thrown, preventing the login.
```
def complete_social_login(request, sociallogin):
assert not sociallogin.is_existing …
sociallogin.lookup()
try:
get_adapter().pre_social_login(request, sociallogin)
signals.pre_social_login.send(
sender=SocialLogin, request=request, sociallogin=sociallogin
)
```
Current Behavior:
- After a new user registers through a social account, attempting to log in with any existing user accounts results in an AssertionError.
Expected Behavior:
- Existing users should be able to log in successfully, regardless of new users registering through social accounts.
Workarounds:
Restarting the Django server temporarily resolves the issue, allowing existing users to log in again without encountering the AssertionError.
Deleting the newly registered user also returns the system to normal, eliminating the login issue for existing users.
I am looking for guidance on how to permanently resolve this issue without resorting to restarting the server or deleting new users. Any insights or suggestions would be greatly appreciated.
Thank you for your assistance.
|
closed
|
2024-03-11T15:33:23Z
|
2025-02-23T07:45:56Z
|
https://github.com/pennersr/django-allauth/issues/3679
|
[] |
YuchenTOR
| 4
|
koxudaxi/datamodel-code-generator
|
pydantic
| 2,130
|
The class "Extra" is deprecated Extra is deprecated. Use literal values instead (e.g. `extra='allow'`)
|
**Describe the bug**
This is not a bug **yet**, but I think I should report it.
So, I just installed `datamodel-code-generator` to be able to convert a JSON schema to a dataclass.
Here's the dependencies
```
datamodel-code-generator 0.26.2
...
pydantic 2.9.2
pydantic_core 2.23.4
```
When I execute the command below, I get several warnings, such as
> The class "Extra" is deprecated
Extra is deprecated. Use literal values instead (e.g. `extra='allow'`)
or
> Call expression not allowed in type expression Pylance [reportInvalidTypeForm](https://github.com/microsoft/pyright/blob/main/docs/configuration.md#reportInvalidTypeForm)
When I try to convert the dataclass back to a JSON schema, I get another similar warning
> PydanticDeprecatedSince20: `pydantic.config.Extra` is deprecated, use literal values instead (e.g. `extra='allow'`). Deprecated in Pydantic V2.0 to be removed in V3.0. See Pydantic V2 Migration Guide at https://errors.pydantic.dev/2.9/migration/
The generated model indeed imports `Extra` and defines the model as follows
```
class Model(BaseModel):
class Config:
extra = Extra.forbid # Extra is deprecated
text: constr(min_length=3, max_length=16) # constr is deprecated
...
```
The "extra" code is generated because I have `"additionalProperties": false` in my schema, while the `constr` is added because I have something like this
```
"text": {
"type": "string",
"minLength": 3,
"maxLength": 16
},
```
Before I proceed to use this library, are you planning to solve these issues?
**To Reproduce**
See example above
Used commandline:
```
datamodel-codegen --input schema.json --input-file-type jsonschema --output model.py
```
**Expected behavior**
No warning
**Version:**
- OS: MacOS Sonoma
- Python version: 3.11.9
- datamodel-code-generator version: 0.26.2
|
closed
|
2024-10-21T15:06:26Z
|
2024-10-22T17:04:00Z
|
https://github.com/koxudaxi/datamodel-code-generator/issues/2130
|
[] |
nbro10
| 2
|
ultralytics/ultralytics
|
pytorch
| 19,310
|
Device selection on export on multi-gpu systems
|
### Search before asking
- [x] I have searched the Ultralytics YOLO [issues](https://github.com/ultralytics/ultralytics/issues) and found no similar bug report.
### Ultralytics YOLO Component
_No response_
### Bug
Greatings! 🚀
Sorry for my English. I ran into issue (latest version, February, 19th) when choosing GPU to export on NVIDIA mGPU setup:
```
DEVICE0 = "cuda:1"
torch.set_default_device(device=DEVICE0)
with torch.cuda.device(device=DEVICE0):
model = YOLO("yolo11m.pt")
model.export(format="engine", half=True, imgsz=TRACK_HW, batch=BATCH_SIZE, dynamic=True, device=DEVICE0)
```
I selected second (:1) gpu, but got a usage on first (:0) one. nvidia-smi showed a full load on first gpu with a small one on second.
utils/torch_utils.py
```
if not cpu and not mps and torch.cuda.is_available(): # prefer GPU if available
devices = device.split(",") if device else "0" # i.e. "0,1" -> ["0", "1"]
n = len(devices) # device count
if n > 1: # multi-GPU
if batch < 1:
raise ValueError(
"AutoBatch with batch<1 not supported for Multi-GPU training, "
"please specify a valid batch size, i.e. batch=16."
)
if batch >= 0 and batch % n != 0: # check batch_size is divisible by device_count
raise ValueError(
f"'batch={batch}' must be a multiple of GPU count {n}. Try 'batch={batch // n * n}' or "
f"'batch={batch // n * n + n}', the nearest batch sizes evenly divisible by {n}."
)
space = " " * (len(s) + 1)
for i, d in enumerate(devices):
s += f"{'' if i == 0 else space}CUDA:{d} ({get_gpu_info(i)})\n" # bytes to MB
arg = "cuda:0"
```
The line leads to bug: ```arg = "cuda:0"```
I suppose it should be: ```arg = f"cuda:{device}"```
### Environment
Package Version
------------------------- ------------
addict 2.4.0
aiohappyeyeballs 2.4.4
aiohttp 3.11.11
aiosignal 1.3.2
albucore 0.0.23
albumentations 2.0.2
annotated-types 0.7.0
anyio 4.8.0
attrs 25.1.0
bcrypt 4.2.1
certifi 2025.1.31
cffi 1.17.1
chardet 5.2.0
charset-normalizer 3.4.1
click 8.1.8
coloredlogs 15.0.1
contourpy 1.3.1
cryptography 44.0.0
cycler 0.12.1
fastapi 0.115.6
filelock 3.17.0
flatbuffers 25.1.24
fonttools 4.55.7
frozenlist 1.5.0
fsspec 2025.2.0
geographiclib 2.0
greenlet 3.1.1
h11 0.14.0
huggingface-hub 0.27.1
humanfriendly 10.0
idna 3.10
Jinja2 3.1.5
jsonschema 4.23.0
jsonschema-specifications 2024.10.1
jwt 1.3.1
kiwisolver 1.4.8
lap 0.5.12
lightning-utilities 0.11.9
MarkupSafe 3.0.2
matplotlib 3.10.0
mpmath 1.3.0
msgpack 1.1.0
multidict 6.1.0
networkx 3.4.2
numpy 2.1.1
nvidia-cublas-cu12 12.6.4.1
nvidia-cuda-cupti-cu12 12.6.80
nvidia-cuda-nvrtc-cu12 12.6.77
nvidia-cuda-runtime-cu12 12.6.77
nvidia-cudnn-cu12 9.5.1.17
nvidia-cufft-cu12 11.3.0.4
nvidia-curand-cu12 10.3.7.77
nvidia-cusolver-cu12 11.7.1.2
nvidia-cusparse-cu12 12.5.4.2
nvidia-cusparselt-cu12 0.6.3
nvidia-nccl-cu12 2.21.5
nvidia-nvjitlink-cu12 12.6.85
nvidia-nvtx-cu12 12.6.77
onnx 1.17.0
onnxruntime-gpu 1.20.1
onnxslim 0.1.48
opencv-python 4.11.0.86
opencv-python-headless 4.11.0.86
openvino 2025.0.0
openvino-telemetry 2025.0.0
packaging 24.2
pandas 2.2.3
pillow 11.1.0
pip 24.3.1
propcache 0.2.1
protobuf 5.29.3
psutil 6.1.1
psycopg2-binary 2.9.10
py-cpuinfo 9.0.0
pyarrow 19.0.0
pycparser 2.22
pydantic 2.10.5
pydantic_core 2.27.2
PyJWT 2.10.1
pyparsing 3.2.1
pysrt 1.1.2
python-dateutil 2.9.0.post0
python-dotenv 1.0.1
python-magic 0.4.27
python-multipart 0.0.20
pytorch-lightning 2.5.0.post0
pytz 2025.1
PyYAML 6.0.2
pyzmq 26.2.0
ray 2.40.0
referencing 0.36.2
requests 2.32.3
rpds-py 0.22.3
safetensors 0.5.2
scipy 1.15.1
seaborn 0.13.2
setuptools 75.8.0
simsimd 6.2.1
six 1.17.0
sniffio 1.3.1
SQLAlchemy 2.0.37
sqlmodel 0.0.22
starlette 0.41.3
stringzilla 3.11.3
sympy 1.13.1
tensorboardX 2.6.2.2
tensorrt 10.7.0.post1
tensorrt_cu12 10.7.0.post1
tensorrt-cu12-bindings 10.7.0.post1
tensorrt-cu12-libs 10.7.0.post1
timm 1.0.14
torch 2.6.0+cu126
torch_tensorrt 2.6.0+cu126
torchaudio 2.6.0+cu126
TorchCodec 0.2.0+cu126
torchmetrics 1.0.3
torchvision 0.21.0+cu126
tqdm 4.67.1
triton 3.2.0
typing_extensions 4.12.2
tzdata 2025.1
ultralytics 8.3.76
ultralytics-thop 2.0.14
urllib3 2.3.0
uvicorn 0.34.0
websockets 14.2
wheel 0.45.1
yarl 1.18.3
### Minimal Reproducible Example
```
DEVICE0 = "cuda:1"
torch.set_default_device(device=DEVICE0)
with torch.cuda.device(device=DEVICE0):
model = YOLO("yolo11m.pt")
model.export(format="engine", half=True, imgsz=TRACK_HW, batch=BATCH_SIZE, dynamic=True, device=DEVICE0)
```
### Additional
_No response_
### Are you willing to submit a PR?
- [ ] Yes I'd like to help by submitting a PR!
|
closed
|
2025-02-19T10:30:55Z
|
2025-02-20T18:39:51Z
|
https://github.com/ultralytics/ultralytics/issues/19310
|
[
"exports"
] |
liwtw
| 4
|
pywinauto/pywinauto
|
automation
| 511
|
Maintaining control identifiers in centralized location
|
When desktop application is very large and has 10k identifiers,then how should one maintain and access then from centralized location (file) in python.
Please give suggestions.
Earlier we were using QTP for automation which has its own utility of object repository... For pywinauto how can we create similar utility or how can we organize objects in one file and access them efficiently in script
|
open
|
2018-06-26T15:42:37Z
|
2024-01-09T07:14:02Z
|
https://github.com/pywinauto/pywinauto/issues/511
|
[
"enhancement",
"Priority-Critical",
"good first issue"
] |
NeJaiswal
| 12
|
FactoryBoy/factory_boy
|
django
| 598
|
Changes introduced in #345 altered how django_get_or_create behaves
|
Upgrading to 2.12.0 broke some of the tests we had, and I'm pretty sure either the docs for django_get_or_create are incorrect or the change in behavior was unintended. The change that seems to be causing the problem is https://github.com/FactoryBoy/factory_boy/pull/345
We have a model with three fields marked as unique=True.
```python
class MyModel(model):
field1 = models.CharField(max_length=128, unique=True)
field2 = models.CharField(max_length=128, unique=True)
field3 = models.CharField(max_length=128, unique=True)
```
We have a factory for creating MyModel in tests:
```python
class MyModelFactory(factory.DjangoModelFactory):
class Meta:
model = MyModel
django_get_or_create = ('field1',)
field1 = factory.LazyAttribute(lambda x: fake.text(max_nb_chars=60))
field2 = factory.LazyAttribute(lambda x: fake.text(max_nb_chars=60))
field3 = factory.LazyAttribute(lambda x: fake.text(max_nb_chars=60))
```
We have only one of the unique fields (field1) included in django_get_or_create. Then we have a different test that creates two MyModels with the same field2, and we expect the second creation to fail with a unique constraint exception.
```python
def test_duplicate_field2_not_allowed(self):
MyModelFactory(field2="something")
with pytest.raises(IntegrityError):
MyModelFactory(field2="something")
```
This works in 2.11.1, and fails in 2.12.0.
It seems from the docs:
```
Fields whose name are passed in this list will be used to perform a Model.objects.get_or_create() instead of the usual Model.objects.create():
```
that if a field isn't included in django_get_or_create then it shouldn't be used in Model.objects.get_or_create() ever.
|
closed
|
2019-05-24T19:20:35Z
|
2019-05-28T14:38:48Z
|
https://github.com/FactoryBoy/factory_boy/issues/598
|
[
"Bug",
"Django"
] |
mkokotovich
| 1
|
encode/httpx
|
asyncio
| 3,334
|
Document `Authentication` header is stripped on redirection
|
- [x] Initially raised as discussion #3291
|
open
|
2024-10-08T01:44:02Z
|
2024-10-28T21:27:39Z
|
https://github.com/encode/httpx/issues/3334
|
[
"docs"
] |
findmyway
| 0
|
google-research/bert
|
nlp
| 674
|
Not compatible with tensorflow 2.0
|
Is Bert not compatible with tensorflow 2.0 ?
AttributeError Traceback (most recent call last)
<ipython-input-4-1b957e7a053a> in <module>()
1 import modeling
----> 2 import optimization
3 import run_classifier
4 import run_classifier_with_tfhub
5 import tokenization
/content/bert_repo/optimization.py in <module>()
85
86
---> 87 class AdamWeightDecayOptimizer(tf.train.Optimizer):
88 """A basic Adam optimizer that includes "correct" L2 weight decay."""
89
AttributeError: module 'tensorflow._api.v2.train' has no attribute 'Optimizer'
|
closed
|
2019-06-04T05:55:20Z
|
2020-09-02T09:56:05Z
|
https://github.com/google-research/bert/issues/674
|
[] |
makaveli10
| 5
|
CorentinJ/Real-Time-Voice-Cloning
|
python
| 816
|
Is this supposed to happen? am I supposed to put an audio file next to this?
|
Hello, I get this error in Ubuntu on my WSL 2 implementation and Ubuntu Docker NOVNC container. Please place more details or a pathway in documentation to help with error resolution
See below
############
**gitpod /workspace/Real-Time-Voice-Cloning $** python demo_cli.py
Arguments:
enc_model_fpath: encoder/saved_models/pretrained.pt
syn_model_fpath: synthesizer/saved_models/pretrained/pretrained.pt
voc_model_fpath: vocoder/saved_models/pretrained/pretrained.pt
cpu: False
no_sound: False
seed: None
no_mp3_support: False
Traceback (most recent call last):
File "demo_cli.py", line 42, in <module>
import sounddevice as sd
File "/workspace/.pip-modules/lib/python3.8/site-packages/sounddevice.py", line 71, in <module>
raise OSError('PortAudio library not found')
OSError: PortAudio library not found
**gitpod /workspace/Real-Time-Voice-Cloning $**
###########
|
closed
|
2021-08-11T20:51:14Z
|
2021-08-25T08:48:55Z
|
https://github.com/CorentinJ/Real-Time-Voice-Cloning/issues/816
|
[] |
Hi-ip
| 1
|
akfamily/akshare
|
data-science
| 5,227
|
AKShare 接口问题报告 - stock_board_industry_index_ths
|
**详细问题描述**
1. 请先详细阅读文档对应接口的使用方式:https://akshare.akfamily.xyz
2. 操作系统版本:Win11 64位
3. Python 版本:3.8 以上的版本
4. AKShare 版本:最新版
5. 接口的名称和相应的调用代码
接口名称:stock_board_industry_index_ths
调用:http://127.0.0.1:8080/api/public/stock_board_industry_index_ths?symbol=元件&start_date=20100930&end_date=20241009
7. 接口报错的截图或描述
其他的接口调用没有问题,只有这个接口一直报错:
raise FeatureNotFound(
bs4.FeatureNotFound: Couldn't find a tree builder with the features you requested: lxml. Do you need to install a parser library?
<img width="892" alt="error" src="https://github.com/user-attachments/assets/bb8824da-b24b-4a52-8fb7-ad60abb824f2">
9. 期望获得的正确结果
显示行业数据
|
closed
|
2024-10-07T13:06:43Z
|
2024-10-11T14:11:52Z
|
https://github.com/akfamily/akshare/issues/5227
|
[
"bug"
] |
jiataocai
| 2
|
QingdaoU/OnlineJudge
|
django
| 463
|
纯小白求助
|
请问大家是怎么进行二次开发的呢 我在ubuntu上部署了FE和onlinejudge,在ubuntu上只能看到前端页面,也进不去后台管理,课设需要在青岛oj上开发,不太了解这种架构的开发方式,部署完了,怎么对前端和后端二次开发,有没有对这个项目有过二开经验的大佬,求有偿指导
|
closed
|
2024-04-15T14:15:40Z
|
2024-04-17T08:35:50Z
|
https://github.com/QingdaoU/OnlineJudge/issues/463
|
[] |
t1yifang
| 1
|
strawberry-graphql/strawberry
|
django
| 2,864
|
Problems with from __future__ import annotations and relay
|
<!-- Provide a general summary of the bug in the title above. -->
The novel relay module is not compatible with `from __future__ import annotations`.
I build a simple demo:
https://github.com/devkral/graphene-protector/tree/demo/withresolve_bug (see first section how to reproduce)
Somehow the field cannot be correctly parsed from its string form.
<!-- A clear and concise description of what the bug is. -->
## System Information
- Strawberry version (if applicable): 0.186.1
|
open
|
2023-06-19T09:13:50Z
|
2025-03-20T15:56:15Z
|
https://github.com/strawberry-graphql/strawberry/issues/2864
|
[
"bug"
] |
devkral
| 3
|
paperless-ngx/paperless-ngx
|
django
| 9,192
|
[BUG] Deleting a Split in Multi-Split Documents Deletes the Wrong Split
|
### Description
When splitting documents I ran into this problem where if you have a document with many splits and then try to delete a split, it will fail to delete the selected split.
### Steps to reproduce
1. Download this sample pdf [bla4.pdf](https://github.com/user-attachments/files/18921583/bla4.pdf). Having said this, I think it is possible to reproduce the bug with any document that has many pages.
2. Open the split tool inside Paperless on the big document
3. Create small splits for the first 12 pages that only contain 1 page
4. Delete the Split that contains only page 10
**Expected**:
The split that consists only of page 10 is being deleted and a new split is created that contains pages 10-11
**What actually happens**:
The split that consists of page 7 is being deleted which leads to a new split that consists of pages 7 and 8 being created.
Interestingly everything works fine if you create less splits:
1. Create single page splits for the first 5 pages
2. Delete the split that consists of page 3
What actually happens and also is expected:
The split that consits of page 3 is deleted and a new split ís created that consists of page 3-4.
In this GIF you can see the bug happening:

### Webserver logs
```bash
There is no logs, because splitting hasn't happened yet.
```
### Browser logs
```bash
```
### Paperless-ngx version
2.14.5
### Host OS
Ubuntu 22.04
### Installation method
Docker - official image
### System status
```json
```
### Browser
Chrome
### Configuration changes
_No response_
### Please confirm the following
- [x] I believe this issue is a bug that affects all users of Paperless-ngx, not something specific to my installation.
- [x] This issue is not about the OCR or archive creation of a specific file(s). Otherwise, please see above regarding OCR tools.
- [x] I have already searched for relevant existing issues and discussions before opening this report.
- [x] I have updated the title field above with a concise description.
|
closed
|
2025-02-22T10:34:24Z
|
2025-02-22T15:28:27Z
|
https://github.com/paperless-ngx/paperless-ngx/issues/9192
|
[
"bug",
"frontend"
] |
gooney47
| 2
|
mars-project/mars
|
scikit-learn
| 2,758
|
make mars type inference optional
|
<!--
Thank you for your contribution!
Please review https://github.com/mars-project/mars/blob/master/CONTRIBUTING.rst before opening an issue.
-->
**Is your feature request related to a problem? Please describe.**
Mars type inferrence will generate mock data, then feed the data into user provided function, if the user function is time-consuming which take minutes or hours, there will be much time wasting in type inference, and the real computing didn't even happen which is unacceptable in some cases.
**Describe the solution you'd like**
It would be great if we can make dtypes lazy and type inferrence optional. If so, the expression call cost would be minimal.
|
closed
|
2022-02-25T10:40:07Z
|
2022-02-28T06:15:22Z
|
https://github.com/mars-project/mars/issues/2758
|
[
"type: enhancement",
"mod: dataframe"
] |
chaokunyang
| 0
|
pyeve/eve
|
flask
| 605
|
GridFSMediaStorage does not save filename
|
`GridFSMediaStorage.put` method is saving the filename from the keyword argument, however `store_media_files` in `eve.methods.common` does not specify anything for filename argument.
I have a workaround currently by subclassing `GridFSMediaStorage.put` method but default behavior should also specify the filename for keyword argument
|
closed
|
2015-04-18T02:32:03Z
|
2015-04-18T07:07:23Z
|
https://github.com/pyeve/eve/issues/605
|
[] |
slamuu
| 0
|
sqlalchemy/alembic
|
sqlalchemy
| 369
|
ORM session creates a subtransaction on get_bind()? this interferes w/ things ?
|
**Migrated issue, originally created by chris7 ([@chris7](https://github.com/chris7))**
I am trying to run a trivial migration, and alembic will migrate the database, but is not updating the database version. To update alembic_version, it requires me to run the migration again.
Here's the script I am running:
```python
def upgrade():
op.add_column(
'peaks',
sa.Column('rti', sa.Float(), nullable=True),
)
```
And the output of a full migration:
```
alembic upgrade head
INFO [alembic.runtime.migration] Context impl SQLiteImpl.
INFO [alembic.runtime.migration] Will assume non-transactional DDL.
INFO [alembic.runtime.migration] Running upgrade 1 -> 2, Add peakgroups
INFO [alembic.runtime.migration] Running upgrade 2 -> 3, peakgroup_feature_reference
INFO [alembic.runtime.migration] Running upgrade 3 -> 4, feature_to_peakgroup
INFO [alembic.runtime.migration] Running upgrade 4 -> 5, remove feature peaks
INFO [alembic.runtime.migration] Running upgrade 5 -> 6, add retention time indices
$ alembic current
INFO [alembic.runtime.migration] Context impl SQLiteImpl.
INFO [alembic.runtime.migration] Will assume non-transactional DDL.
5
```
The database at this point has the 'rti' column, so it was successful but the alembic_version table was never updated. I can run the migration again, and it will stamp the database with 6.
running alembic v. 0.8.6 & SQLAlchemy 1.0.12. The database is a sqlite file.
|
closed
|
2016-04-17T15:17:38Z
|
2017-02-23T16:36:39Z
|
https://github.com/sqlalchemy/alembic/issues/369
|
[
"bug"
] |
sqlalchemy-bot
| 10
|
aio-libs/aiomysql
|
sqlalchemy
| 102
|
Connecting via URL instead of separate connection values
|
Could we connect by passing the connection details via an URL instead of having separate host, port, etc. variables? [Similar to what aioamqp has with its from_url function](https://github.com/Polyconseil/aioamqp/blob/b1d11024c658e03722bee57f97a9ced8e3e6b1bc/aioamqp/__init__.py#L76).
|
closed
|
2016-09-01T11:35:57Z
|
2021-10-01T00:18:14Z
|
https://github.com/aio-libs/aiomysql/issues/102
|
[] |
ghost
| 3
|
postmanlabs/httpbin
|
api
| 285
|
test_post_body_unicode fails on PyPy 3 with UnicodeDecodeError
|
PyPy 3 raises an exception during `test_post_body_unicode`
```
RPython traceback:
...
Fatal RPython error: UnicodeDecodeError
```
Is httpbin interested in fixes for PyPy 3?
|
closed
|
2016-05-02T03:46:08Z
|
2018-04-26T17:51:10Z
|
https://github.com/postmanlabs/httpbin/issues/285
|
[] |
jayvdb
| 2
|
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.