repo_name
stringlengths 9
75
| topic
stringclasses 30
values | issue_number
int64 1
203k
| title
stringlengths 1
976
| body
stringlengths 0
254k
| state
stringclasses 2
values | created_at
stringlengths 20
20
| updated_at
stringlengths 20
20
| url
stringlengths 38
105
| labels
listlengths 0
9
| user_login
stringlengths 1
39
| comments_count
int64 0
452
|
|---|---|---|---|---|---|---|---|---|---|---|---|
encode/httpx
|
asyncio
| 2,304
|
unhashable type: 'bytearray'
| ERROR: type should be string, got "\r\nhttps://github.com/encode/httpx/discussions\r\n\r\nGives the error \"unhashable type: 'bytearray' \" when I make a GET request using cookies, headers and parameters\r\n\r\nCode:\r\n`\r\nfor _ in range(retries):\r\n response = await self.session.request(\r\n method=method, url=url, data=data, params=params,headers=headers,\r\n follow_redirects=allow_redirects, timeout=35\r\n )\r\n`"
|
closed
|
2022-07-13T19:39:44Z
|
2022-07-14T12:34:45Z
|
https://github.com/encode/httpx/issues/2304
|
[] |
Vigofs
| 2
|
Kludex/mangum
|
asyncio
| 105
|
`rawQueryString` seems to be a string instead of bytes literals
|
👋 @erm, first let me thank you for this ~great~ awesome library, as the author of another `proxy` for aws lambda I know this can be a 🕳️ sometimes.
I'm currently working on https://github.com/developmentseed/titiler which is a FastAPI based tile server. When deploying the lambda function using mangum and the new api gateway HTTP endpoint, most of my endpoints works except for the ones where I call `request.url.scheme`
e.g: https://github.com/developmentseed/titiler/blob/master/titiler/api/api_v1/endpoints/metadata.py#L59
and I get `AttributeError: 'str' object has no attribute 'decode'`
#### Logs
```
File "/tmp/pip-unpacked-wheel-a0vaawkp/titiler/api/api_v1/endpoints/metadata.py", line 59, in tilejson
File "/tmp/pip-unpacked-wheel-xa0z_c8l/starlette/requests.py", line 56, in url
File "/tmp/pip-unpacked-wheel-xa0z_c8l/starlette/datastructures.py", line 45, in __init__
AttributeError: 'str' object has no attribute 'decode'
```
So looking at my cloudwatch logs I can deduce:
1. in https://github.com/encode/starlette/blob/97257515f8806b8cc519d9850ecadd783b3008f9/starlette/datastructures.py#L45 starlette expect the `query_string` to be a byte literal
2. in https://github.com/erm/mangum/blob/4fbf1b0d7622c19385a549eddfb603399a125bbb/mangum/adapter.py#L100-L106
it seems that Mangum is getting the `rawQueryString` for the event, and in my case I think this would be a string instead of a byte literal.
I'll work on adding logging and test in my app to confirm this, but I wanted to share this to see if I was making sense ;-)
Thanks
|
closed
|
2020-05-09T02:55:34Z
|
2020-05-09T14:23:05Z
|
https://github.com/Kludex/mangum/issues/105
|
[] |
vincentsarago
| 5
|
litestar-org/litestar
|
pydantic
| 3,758
|
Enhancement: "Remember me" support for ServerSideSessionBackend / ClientSideSessionBackend
|
### Summary
"Remember me" checkbox during user log in is a common practice currently does not supported by implementations of ServerSideSessionBackend / ClientSideSessionBackend classes.
### Basic Example
```
def set_session(self, value: dict[str, Any] | DataContainerType | EmptyType) -> None:
"""Set the session in the connection's ``Scope``.
If the :class:`SessionMiddleware <.middleware.session.base.SessionMiddleware>` is enabled, the session will be added
to the response as a cookie header.
Args:
value: Dictionary or pydantic model instance for the session data.
Returns:
None
"""
self.scope["session"] = value
```
I think the best way would be to add ability to set specific cookie params in 'set_session' method, they would be used over default ones specified in SessionAuth instance. Then add support for it for ServerSideSessionBackend / ClientSideSessionBackend.
### Drawbacks and Impact
Need to be careful about backward compability but i think there should not be much of a problem.
### Unresolved questions
_No response_
|
open
|
2024-09-25T07:26:20Z
|
2025-03-20T15:54:55Z
|
https://github.com/litestar-org/litestar/issues/3758
|
[
"Enhancement"
] |
Rey092
| 0
|
ivy-llc/ivy
|
tensorflow
| 28,648
|
Fix Frontend Failing Test: jax - averages_and_variances.numpy.average
|
To-do List: https://github.com/unifyai/ivy/issues/27496
|
closed
|
2024-03-19T18:21:46Z
|
2024-03-26T04:48:36Z
|
https://github.com/ivy-llc/ivy/issues/28648
|
[
"Sub Task"
] |
ZJay07
| 0
|
fastapi-users/fastapi-users
|
fastapi
| 600
|
Backref not working at route users/me
|
**Using fastapi-users 3.0.6 and tortoise orm 0.16.17**
I have been stuck by almost a week trying to solve this problem and now it seems to be a bug, so I'm leaving evidence.
I want to access the route **users/me** and append some extra data from a couple of relations I created, these are:
**Tortoise models**
class UserModel(TortoiseBaseUserModel):
nombre = fields.CharField(max_length=100)
apellidos = fields.CharField(max_length=100)
fecha_nacimiento = fields.DateField(default=None)
telefono = fields.CharField(max_length=20)
rut = fields.CharField(max_length=10)
cargo = fields.CharField(max_length=30)
class Rol(models.Model):
id = fields.UUIDField(pk=True)
nombre = fields.CharField(max_length=100)
class Permiso(models.Model):
id = fields.UUIDField(pk=True)
nombre = fields.CharField(max_length=100)
class Auth_Rol_Permiso(models.Model, TimestampMixin): -->Relational table between user,permiso and rol classes
id = fields.UUIDField(pk=True)
rol = fields.ForeignKeyField('models.Rol', related_name="auth_rol", on_delete=fields.CASCADE)
permiso = fields.ForeignKeyField('models.Permiso', related_name="auth_permiso", on_delete=fields.CASCADE)
user = fields.ForeignKeyField("models.UserModel", related_name="auth_user", on_delete=fields.CASCADE)
So, for Pydantic User schema, I created this:
class User(fastapi_users_model.BaseUser):
nombre: str
apellidos: str
fecha_nacimiento: date
telefono: str
rut: str
cargo: str
auth_user: List[UserPermission] --> Backref for getting permiso and rol relation data
class Permisoo(BaseModel):
id: UUID
nombre: str
class UserPermission(BaseModel):
permiso: Optional[Permisoo]
created_at: datetime
rol: Optional[Dict[str, Any]]
The problem comes when I try to get data from route **users/me**, this is the response:
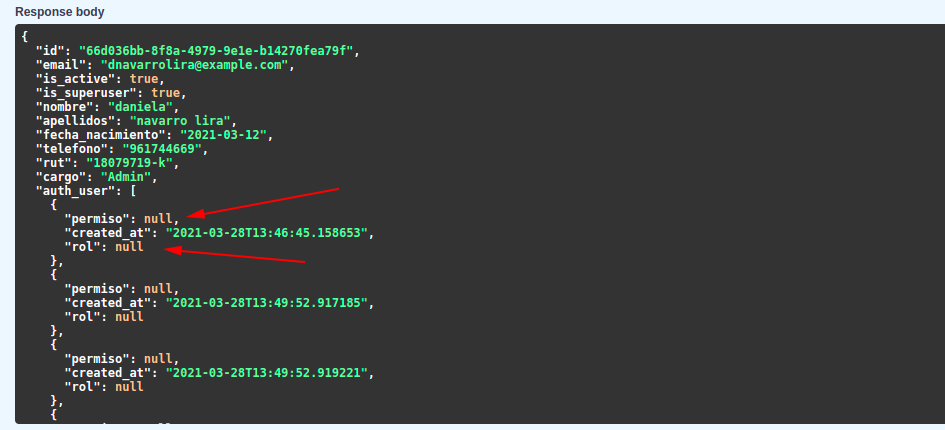
But if I go into a custom endpoint that I create for getting rol class, it's working and getting all data without problems!!...I think maybe the problem is that rol pydantic schema is generated by tortoise, with this function -> **pydantic_model_creator**, like this:
`rol_out = pydantic_model_creator(Rol, name="RolOut")`
Anyway, this is the result of getting rol class(working fine):
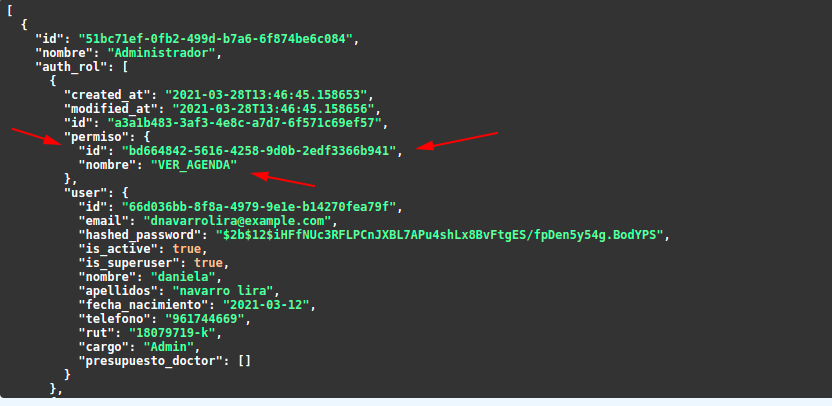
this doesn't make any sense to me, so I hope you can help me, thanks for your time.
|
closed
|
2021-04-12T17:08:12Z
|
2021-04-20T15:06:07Z
|
https://github.com/fastapi-users/fastapi-users/issues/600
|
[
"bug"
] |
Master-Y0da
| 5
|
coqui-ai/TTS
|
python
| 3,530
|
Add "host" argument to the server.py script
|
Hello,
I tried using the default server.py script as-is but it is trying to use the IPv6 by default and I get stuck as I don't have IPv6 in my environment.
The command:
```bash
python3 TTS/server/server.py --model_name tts_models/en/ljspeech/tacotron2-DCA
```
Gives the following error:
```bash
Traceback (most recent call last): │
│ File "/root/TTS/server/server.py", line 258, in <module> │
│ main() │
│ File "/root/TTS/server/server.py", line 254, in main │
│ app.run(debug=args.debug, host="::", port=args.port) │
│ File "/usr/local/lib/python3.10/site-packages/flask/app.py", line 612, in run │
│ run_simple(t.cast(str, host), port, self, **options) │
│ File "/usr/local/lib/python3.10/site-packages/werkzeug/serving.py", line 1077, in run_simple │
│ srv = make_server( │
│ File "/usr/local/lib/python3.10/site-packages/werkzeug/serving.py", line 917, in make_server │
│ return ThreadedWSGIServer( │
│ File "/usr/local/lib/python3.10/site-packages/werkzeug/serving.py", line 737, in __init__ │
│ super().__init__( │
│ File "/usr/local/lib/python3.10/socketserver.py", line 448, in __init__ │
│ self.socket = socket.socket(self.address_family, │
│ File "/usr/local/lib/python3.10/socket.py", line 232, in __init__ │
│ _socket.socket.__init__(self, family, type, proto, fileno) │
│ OSError: [Errno 97] Address family not supported by protocol
```
|
closed
|
2024-01-19T13:04:43Z
|
2024-03-02T00:44:46Z
|
https://github.com/coqui-ai/TTS/issues/3530
|
[
"wontfix",
"feature request"
] |
spartan
| 2
|
gee-community/geemap
|
jupyter
| 586
|
ValueError: Unknown color None. when trying to use a coloramp
|
<!-- Please search existing issues to avoid creating duplicates. -->
### Environment Information
- geemap version: 0.8.18
- Python version: 3
- Operating System: google collab
### Description
```
---------------------------------------------------------------------------
ValueError Traceback (most recent call last)
<ipython-input-8-8010224dd26d> in <module>()
4 vmax = visPara['max']
5
----> 6 Map.add_colorbar(visPara, label="Elevation (m a.s.l)", orientation="vertical", layer_name="Arctic DEM")
7 # Map.add_colorbar_branca(colors=colors, vmin=vmin, vmax=vmax, caption="m a.s.l")
3 frames
/usr/local/lib/python3.7/dist-packages/branca/colormap.py in _parse_color(x)
43 cname = _cnames.get(x.lower(), None)
44 if cname is None:
---> 45 raise ValueError('Unknown color {!r}.'.format(cname))
46 color_tuple = _parse_hex(cname)
47 else:
ValueError: Unknown color None.
```
### What I Did
For either my geemap code or even this example (https://colab.research.google.com/github/fsn1995/GIS-at-deep-purple/blob/main/02%20gee%20map%20greenland.ipynb#scrollTo=RJdNLlhQjajw) I cant get the colorbars to display (see error above). On the other hand using the same version of geemap etc on my own computer in a Jupyter notebook running in conda it works. It seems to be some issue when I try to run the exact same code in google collab; or that's a red herring and I am doing something else wrong
Here is the code in that example that fails
```
greenlandmask = ee.Image('OSU/GIMP/2000_ICE_OCEAN_MASK') \
.select('ice_mask').eq(1); #'ice_mask', 'ocean_mask'
arcticDEM = ee.Image('UMN/PGC/ArcticDEM/V3/2m_mosaic')
arcticDEMgreenland = arcticDEM.updateMask(greenlandmask)
palette = cm.get_palette('terrain', n_class=10)
visPara = {'min': 0, 'max': 2500.0, 'palette': ['0d13d8', '60e1ff', 'ffffff']}
visPara
# visPara = {'min': 0, 'max': 2500.0, 'palette': palette}
Map.addLayer(arcticDEMgreenland, visPara, 'Arctic DEM')
Map.setCenter(-41.0, 74.0, 3)
#add colorbar
colors = visPara['palette']
vmin = visPara['min']
vmax = visPara['max']
Map.add_colorbar(visPara, label="Elevation (m a.s.l)", orientation="vertical", layer_name="Arctic DEM")
# Map.add_colorbar_branca(colors=colors, vmin=vmin, vmax=vmax, caption="m a.s.l")
```
or
```
greenlandmask = ee.Image('OSU/GIMP/2000_ICE_OCEAN_MASK') \
.select('ice_mask').eq(1); #'ice_mask', 'ocean_mask'
arcticDEM = ee.Image('UMN/PGC/ArcticDEM/V3/2m_mosaic')
arcticDEMgreenland = arcticDEM.updateMask(greenlandmask)
palette = cm.get_palette('terrain', n_class=10)
# visPara = {'min': 0, 'max': 2500.0, 'palette': ['0d13d8', '60e1ff', 'ffffff']}
visPara = {'min': 0, 'max': 2500.0, 'palette': palette}
Map.addLayer(arcticDEMgreenland, visPara, 'Arctic DEM terrain')
Map.setCenter(-41.0, 74.0, 3)
Map.add_colorbar(visPara, label="Elevation (m)", discrete=False, orientation="vertical", layer_name="Arctic DEM terrain")
```
or in my code (again works on my computer in Jupyter)
```
### Get GEE imagery
# DEM
elev_dataset = ee.Image('USGS/NED')
ned_elevation = elev_dataset.select('elevation')
#colors = cmr.take_cmap_colors('viridis', 5, return_fmt='hex')
#colors = cmr.take_cmap_colors('viridis', None, cmap_range=(0.2, 0.8), return_fmt='hex')
#dem_palette = cmr.take_cmap_colors(mpl.cm.get_cmap('Spectral', 30).reversed(),30, return_fmt='hex')
dem_palette = cm.get_palette('Spectral_r', n_class=30)
demViz = {'min': 0.0, 'max': 2000.0, 'palette': dem_palette, 'opacity': 1}
# LandFire
lf = ee.ImageCollection('LANDFIRE/Vegetation/EVH/v1_4_0');
lf_evh = lf.select('EVH')
#evh_palette = cmr.take_cmap_colors(mpl.cm.get_cmap('Spectral', 30).reversed(),30, return_fmt='hex')
evh_palette = cm.get_palette('Spectral_r', n_class=30)
evhViz = {'min': 0.0, 'max': 30.0, 'palette': evh_palette, 'opacity': 1}
# Global Forest Canopy Height (2005)
gfch = ee.Image('NASA/JPL/global_forest_canopy_height_2005');
forestCanopyHeight = gfch.select('1');
#gfch_palette = cmr.take_cmap_colors(mpl.cm.get_cmap('Spectral', 30).reversed(),30, return_fmt='hex')
gfch_palette = cm.get_palette('Spectral_r', n_class=30)
gfchViz = {'min': 0.0, 'max': 40.0, 'palette': gfch_palette, 'opacity': 1}
### Plot the elevation data
# Prints the elevation of Mount Everest.
xy = ee.Geometry.Point(sitecoords[:2])
site_elev = ned_elevation.sample(xy, 30).first().get('elevation').getInfo()
print('Selected site elevation (m):', site_elev)
# display NED data
# https://geemap.org/notebooks/14_legends/
# https://geemap.org/notebooks/49_colorbar/
# https://colab.research.google.com/github/fsn1995/GIS-at-deep-purple/blob/main/02%20gee%20map%20greenland.ipynb#scrollTo=uwLvDKHDlN0l
#colors = demViz['palette']
#vmin = demViz['min']
#vmax = demViz['max']
srtm = geemap.Map(center=[33.315809,-85.198609], zoom=6)
srtm.addLayer(ned_elevation, demViz, 'Elevation above sea level');
srtm.addLayer(xy, {'color': 'black', 'strokeWidth': 1}, 'Selected Site')
#srtm.add_colorbar_branca(colors=colors, vmin=vmin, vmax=vmax, label="Elevation (m)",
# orientation="vertical", layer_name="SRTM DEM")
srtm.add_colorbar(demViz, label="Elevation (m)", layer_name="SRTM DEM")
states = ee.FeatureCollection('TIGER/2018/States')
statesImage = ee.Image().paint(states, 0, 2)
srtm.addLayer(statesImage, {'palette': 'black'}, 'US States')
srtm.addLayerControl()
srtm
```
here is the script on GitHub
https://github.com/serbinsh/amf3_seus/blob/main/python/amf3_radar_blockage_demo_gee.ipynb
Any ideas? I was using a different colormap system (cmasher) that also worked for me but not on collab. I switched to the geemap version based on that geemap lib example in the hopes it would fix this "none" issue but it doesn't seem to have fixed it. I wonder if its a geemap versioning issue?
|
closed
|
2021-07-16T16:26:55Z
|
2021-07-19T15:06:05Z
|
https://github.com/gee-community/geemap/issues/586
|
[
"bug"
] |
serbinsh
| 7
|
PaddlePaddle/models
|
computer-vision
| 4,740
|
关于se+resnet vd的问题
|
请问 re_resnet_vd中我找到的是这个位置。先经过一个2*2的池化 ,再经过一个1*1的卷积 与原来的131结构add。可以解释一下这样做的原因么,有什么可解释性么? 为什么不能用1*1卷积 s=2 完成2*2池化+1*1卷积呢 谢谢
https://github.com/PaddlePaddle/models/blob/365fe58a0afdfd038350a718e92684503918900b/PaddleCV/image_classification/models/se_resnet_vd.py#L145
|
open
|
2020-07-05T23:54:34Z
|
2024-02-26T05:11:03Z
|
https://github.com/PaddlePaddle/models/issues/4740
|
[] |
lxk767363331
| 2
|
horovod/horovod
|
deep-learning
| 3,958
|
Horovod docker unable to distribute training on another node. Shows error - No module named horovod.runner
|
**Environment:**
1. Framework: (TensorFlow, Keras, PyTorch, MXNet) TensorFlow
2. Framework version: 2.9.2
3. Horovod version: 0.28.1
4. MPI version:4.1.4
5. CUDA version:
6. NCCL version:
7. Python version: 3.8.10
8. Spark / PySpark version: 3.3.0
9. Ray version: 2.5.0
10. OS and version:
11. GCC version:9.4.0
12. CMake version:3.16.3
**Checklist:**
1. Did you search issues to find if somebody asked this question before?
2. If your question is about hang, did you read [this doc](https://github.com/horovod/horovod/blob/master/docs/running.rst)?
3. If your question is about docker, did you read [this doc](https://github.com/horovod/horovod/blob/master/docs/docker.rst)?
4. Did you check if you question is answered in the [troubleshooting guide](https://github.com/horovod/horovod/blob/master/docs/troubleshooting.rst)?
**Bug report:**
I am using the docker image of horovod that I pulled from dockerhub (docker pull horovod/horovod:latest). My setup is that I have two different nodes assigned on HPC. I wanted to dsitribute the training on both nodes and initially I logged in to node1 and ran
`dockerun -np 2 address_of_node2:2 python script_name` and the output was
```
$ horovodrun -np 2 -H xxxx.iitd.ac.in:2 python tensorflow2_mnist.py
Launching horovod task function was not successful:
Attaching 25746 to akshay.cstaff
/etc/profile.d/lang.sh: line 19: warning: setlocale: LC_CTYPE: cannot change locale (C.UTF-8)
/usr/bin/python: No module named horovod.runner
```
Then i ran python command to see if horovd.runner was missing
```
$ python
Python 3.8.10 (default, May 26 2023, 14:05:08)
[GCC 9.4.0] on linux
Type "help", "copyright", "credits" or "license" for more information.
>>> import horovod
>>> horovod.runner
<module 'horovod.runner' from '/usr/local/lib/python3.8/dist-packages/horovod/runner/__init__.py'>
>>>
```
As you can see it is available.
How to proceed from here?
Both my machines are setup for passwordless ssh too.
|
open
|
2023-07-11T11:23:35Z
|
2023-07-11T11:23:35Z
|
https://github.com/horovod/horovod/issues/3958
|
[
"bug"
] |
AkshayRoyal
| 0
|
koxudaxi/datamodel-code-generator
|
pydantic
| 1,996
|
Support NaiveDateTime
|
**Is your feature request related to a problem? Please describe.**
Add support to choose between `AwareDateTime`, `NaiveDateTime` or generic `datetime`.
**Describe the solution you'd like**
A CLI option to choose between the both.
**Describe alternatives you've considered**
Updating the generated models
**Additional context**
Migrating to pydantic v2 has proven to be very time consuming. Typing becomes more strict, which is a good thing, but going the extra mile as to update the full codebase to ensure all `datetime` objects are using a timezone is too demanding when interact with other tools such as SQLAlchemy, etc.
Pydantic V2 supports:
- `AwareDateTime`
- `NaiveDateTime`
- or the more generic `datetime`
Being able to use the later would make things much easier.
Thanks
|
closed
|
2024-06-06T14:56:33Z
|
2024-10-12T16:42:30Z
|
https://github.com/koxudaxi/datamodel-code-generator/issues/1996
|
[] |
pmbrull
| 1
|
mckinsey/vizro
|
pydantic
| 723
|
Fix capitalisation of `Jupyter notebook` to `Jupyter Notebook`
|
- Do a quick search and find for any `Jupyter notebook` version and replace with `Jupyter Notebook` where suitable
|
closed
|
2024-09-19T15:22:09Z
|
2024-10-07T10:16:16Z
|
https://github.com/mckinsey/vizro/issues/723
|
[
"Good first issue :baby_chick:"
] |
huong-li-nguyen
| 3
|
jonaswinkler/paperless-ng
|
django
| 259
|
Post-Hook - How to set ASN
|
Every scanned Document which is archived as paper get's a Number (Always 6-digits, the first 2 Digits are a long time 0 ;-)). This would be in the File-Content after Consumption and OCR by Paperless. Now i would like to extract the number by RegEx 00\d{4} and setting the ASN of the consumed document. I did not find any solution using the manage.py Script.
Any Idea how to handle this? Is it possible to use a self developed SQL-Statement?
Thx.
|
closed
|
2021-01-03T01:41:59Z
|
2021-01-27T10:38:51Z
|
https://github.com/jonaswinkler/paperless-ng/issues/259
|
[] |
andbez
| 13
|
napari/napari
|
numpy
| 7,702
|
[test-bot] pip install --pre is failing
|
The --pre Test workflow failed on 2025-03-15 00:43 UTC
The most recent failing test was on windows-latest py3.12 pyqt6
with commit: cb6f6e6157990806a53f1c58e31e9e7aa4a4966e
Full run: https://github.com/napari/napari/actions/runs/13867508726
(This post will be updated if another test fails, as long as this issue remains open.)
|
closed
|
2025-03-15T00:43:01Z
|
2025-03-15T03:24:08Z
|
https://github.com/napari/napari/issues/7702
|
[
"bug"
] |
github-actions[bot]
| 1
|
lepture/authlib
|
django
| 365
|
Flask client still relies on requests (missing dependecy)
|
**Describe the bug**
I was following this documentation: https://docs.authlib.org/en/latest/client/flask.html
In Installation doc section (https://docs.authlib.org/en/latest/basic/install.html) it says to use Authlib with Flask we just need to install Authlib and Flask.
However, when using `authlib.integrations.flask_client.OAuth` we still get a missing requests dependency. The docs rightfully say that requests is an optional dependency. But the documentation implies (at least to me) that Authlib is able to use `flask.request`, and that `requests` library is not needed if Flask is used.
I'm using `Flask == 2.0.1`
**Error Stacks**
```
webservice_1 | from authlib.integrations.flask_client import OAuth
webservice_1 | File "/usr/local/lib/python3.9/site-packages/authlib/integrations/flask_client/__init__.py", line 3, in <module>
webservice_1 | from .oauth_registry import OAuth
webservice_1 | File "/usr/local/lib/python3.9/site-packages/authlib/integrations/flask_client/oauth_registry.py", line 4, in <module>
webservice_1 | from .integration import FlaskIntegration
webservice_1 | File "/usr/local/lib/python3.9/site-packages/authlib/integrations/flask_client/integration.py", line 4, in <module>
webservice_1 | from ..requests_client import OAuth1Session, OAuth2Session
webservice_1 | File "/usr/local/lib/python3.9/site-packages/authlib/integrations/requests_client/__init__.py", line 1, in <module>
webservice_1 | from .oauth1_session import OAuth1Session, OAuth1Auth
webservice_1 | File "/usr/local/lib/python3.9/site-packages/authlib/integrations/requests_client/oauth1_session.py", line 2, in <module>
webservice_1 | from requests import Session
webservice_1 | ModuleNotFoundError: No module named 'requests'
```
**To Reproduce**
- pip install Authlib Flask
- Follow https://docs.authlib.org/en/latest/client/flask.html
**Expected behavior**
- Authlib to use Flask requests interface, instead of requiring requests.
**Environment:**
- OS: Docker (Apline)
- Python Version: `3.9`
- Authlib Version: `==0.15.4`
**Additional context**
- Could this be because some internal change to Flask 2.0, recently released?
- Could a quick solution be just to update documentation to include requests also for a Flask installation?
|
closed
|
2021-07-16T11:07:18Z
|
2021-07-17T03:16:51Z
|
https://github.com/lepture/authlib/issues/365
|
[
"bug"
] |
sergioisidoro
| 1
|
Evil0ctal/Douyin_TikTok_Download_API
|
web-scraping
| 61
|
求赐教!如何获取用户的总粉丝量(tiktok)
|
您好,非常想请教一下调用哪些方法可以获取到user的followers amount,感谢
|
closed
|
2022-08-06T19:40:03Z
|
2024-11-13T08:19:31Z
|
https://github.com/Evil0ctal/Douyin_TikTok_Download_API/issues/61
|
[
"help wanted"
] |
Ang-Gao
| 6
|
dmlc/gluon-nlp
|
numpy
| 1,526
|
Pre-training scripts should allow resuming from checkpoints
|
## Description
Currently, the ELECTRA pre-training script can't resume from last checkpoint. It will be useful to enhance the script to resume from checkpoints.
|
open
|
2021-02-21T18:23:04Z
|
2021-03-12T18:44:59Z
|
https://github.com/dmlc/gluon-nlp/issues/1526
|
[
"enhancement"
] |
szha
| 3
|
tensorflow/datasets
|
numpy
| 4,874
|
flower
|
* Name of dataset: <name>
* URL of dataset: <url>
* License of dataset: <license type>
* Short description of dataset and use case(s): <description>
Folks who would also like to see this dataset in `tensorflow/datasets`, please thumbs-up so the developers can know which requests to prioritize.
And if you'd like to contribute the dataset (thank you!), see our [guide to adding a dataset](https://github.com/tensorflow/datasets/blob/master/docs/add_dataset.md).
|
closed
|
2023-04-17T09:33:21Z
|
2023-04-18T11:18:04Z
|
https://github.com/tensorflow/datasets/issues/4874
|
[
"dataset request"
] |
y133977
| 1
|
JaidedAI/EasyOCR
|
pytorch
| 1,083
|
Jupyter kernel dies every time I try to use easyocr following a youtube tutorial's Kaggle notebook
|
I was trying locally so just did ! pip install easyocr.
My notebook as a gist: https://gist.github.com/nyck33/9e02014a9b173071ae3dc62fa631454c
The kernel dies every time in the cell that is : `results = reader.readtext(handwriting[2])`
Just tried again and get
```
OutOfMemoryError: CUDA out of memory. Tried to allocate 1.17 GiB (GPU 0; 4.00 GiB total capacity; 1.32 GiB already allocated; 0 bytes free; 1.33 GiB reserved in total by PyTorch) If reserved memory is >> allocated memory try setting max_split_size_mb to avoid fragmentation. See documentation for Memory Management and PYTORCH_CUDA_ALLOC_CONF
```
Mine is a GeForce GTX 1650 on notebook, should have 4GB or the GPU RAM.
Do I really need to dig in and make changes to Pytorch configurations?
conda list shows:
```
# packages in environment at /home/nobu/miniconda3/envs/kerasocr:
#
# Name Version Build Channel
_libgcc_mutex 0.1 main
_openmp_mutex 5.1 1_gnu
absl-py 1.4.0 pypi_0 pypi
anyio 3.7.1 pypi_0 pypi
argon2-cffi 21.3.0 pypi_0 pypi
argon2-cffi-bindings 21.2.0 pypi_0 pypi
arrow 1.2.3 pypi_0 pypi
asttokens 2.2.1 pypi_0 pypi
astunparse 1.6.3 pypi_0 pypi
async-lru 2.0.3 pypi_0 pypi
attrs 23.1.0 pypi_0 pypi
babel 2.12.1 pypi_0 pypi
backcall 0.2.0 pypi_0 pypi
beautifulsoup4 4.12.2 pypi_0 pypi
bleach 6.0.0 pypi_0 pypi
ca-certificates 2023.05.30 h06a4308_0
cachetools 5.3.1 pypi_0 pypi
certifi 2023.5.7 pypi_0 pypi
cffi 1.15.1 pypi_0 pypi
charset-normalizer 3.2.0 pypi_0 pypi
cmake 3.26.4 pypi_0 pypi
comm 0.1.3 pypi_0 pypi
contourpy 1.1.0 pypi_0 pypi
cycler 0.11.0 pypi_0 pypi
debugpy 1.6.7 pypi_0 pypi
decorator 5.1.1 pypi_0 pypi
defusedxml 0.7.1 pypi_0 pypi
easyocr 1.7.0 pypi_0 pypi
editdistance 0.6.2 pypi_0 pypi
efficientnet 1.0.0 pypi_0 pypi
essential-generators 1.0 pypi_0 pypi
exceptiongroup 1.1.2 pypi_0 pypi
executing 1.2.0 pypi_0 pypi
fastjsonschema 2.17.1 pypi_0 pypi
filelock 3.12.2 pypi_0 pypi
flatbuffers 23.5.26 pypi_0 pypi
fonttools 4.41.0 pypi_0 pypi
fqdn 1.5.1 pypi_0 pypi
gast 0.4.0 pypi_0 pypi
google-auth 2.22.0 pypi_0 pypi
google-auth-oauthlib 1.0.0 pypi_0 pypi
google-pasta 0.2.0 pypi_0 pypi
grpcio 1.56.0 pypi_0 pypi
h5py 3.9.0 pypi_0 pypi
idna 3.4 pypi_0 pypi
imageio 2.31.1 pypi_0 pypi
imgaug 0.4.0 pypi_0 pypi
importlib-metadata 6.8.0 pypi_0 pypi
importlib-resources 6.0.0 pypi_0 pypi
ipykernel 6.24.0 pypi_0 pypi
ipython 8.12.2 pypi_0 pypi
isoduration 20.11.0 pypi_0 pypi
jedi 0.18.2 pypi_0 pypi
jinja2 3.1.2 pypi_0 pypi
json5 0.9.14 pypi_0 pypi
jsonpointer 2.4 pypi_0 pypi
jsonschema 4.18.3 pypi_0 pypi
jsonschema-specifications 2023.6.1 pypi_0 pypi
jupyter-client 8.3.0 pypi_0 pypi
jupyter-core 5.3.1 pypi_0 pypi
jupyter-events 0.6.3 pypi_0 pypi
jupyter-lsp 2.2.0 pypi_0 pypi
jupyter-server 2.7.0 pypi_0 pypi
jupyter-server-terminals 0.4.4 pypi_0 pypi
jupyterlab 4.0.3 pypi_0 pypi
jupyterlab-pygments 0.2.2 pypi_0 pypi
jupyterlab-server 2.23.0 pypi_0 pypi
keras 2.13.1 pypi_0 pypi
keras-applications 1.0.8 pypi_0 pypi
keras-ocr 0.9.2 pypi_0 pypi
kiwisolver 1.4.4 pypi_0 pypi
lazy-loader 0.3 pypi_0 pypi
ld_impl_linux-64 2.38 h1181459_1
libclang 16.0.0 pypi_0 pypi
libffi 3.4.4 h6a678d5_0
libgcc-ng 11.2.0 h1234567_1
libgomp 11.2.0 h1234567_1
libstdcxx-ng 11.2.0 h1234567_1
lit 16.0.6 pypi_0 pypi
markdown 3.4.3 pypi_0 pypi
markupsafe 2.1.3 pypi_0 pypi
matplotlib 3.7.2 pypi_0 pypi
matplotlib-inline 0.1.6 pypi_0 pypi
mistune 3.0.1 pypi_0 pypi
mpmath 1.3.0 pypi_0 pypi
nbclient 0.8.0 pypi_0 pypi
nbconvert 7.6.0 pypi_0 pypi
nbformat 5.9.1 pypi_0 pypi
ncurses 6.4 h6a678d5_0
nest-asyncio 1.5.6 pypi_0 pypi
networkx 3.1 pypi_0 pypi
ninja 1.11.1 pypi_0 pypi
notebook-shim 0.2.3 pypi_0 pypi
numpy 1.24.3 pypi_0 pypi
nvidia-cublas-cu11 11.10.3.66 pypi_0 pypi
nvidia-cuda-cupti-cu11 11.7.101 pypi_0 pypi
nvidia-cuda-nvrtc-cu11 11.7.99 pypi_0 pypi
nvidia-cuda-runtime-cu11 11.7.99 pypi_0 pypi
nvidia-cudnn-cu11 8.5.0.96 pypi_0 pypi
nvidia-cufft-cu11 10.9.0.58 pypi_0 pypi
nvidia-curand-cu11 10.2.10.91 pypi_0 pypi
nvidia-cusolver-cu11 11.4.0.1 pypi_0 pypi
nvidia-cusparse-cu11 11.7.4.91 pypi_0 pypi
nvidia-nccl-cu11 2.14.3 pypi_0 pypi
nvidia-nvtx-cu11 11.7.91 pypi_0 pypi
oauthlib 3.2.2 pypi_0 pypi
opencv-python 4.8.0.74 pypi_0 pypi
opencv-python-headless 4.8.0.74 pypi_0 pypi
openssl 3.0.9 h7f8727e_0
opt-einsum 3.3.0 pypi_0 pypi
overrides 7.3.1 pypi_0 pypi
packaging 23.1 pypi_0 pypi
pandas 2.0.3 pypi_0 pypi
pandocfilters 1.5.0 pypi_0 pypi
parso 0.8.3 pypi_0 pypi
pexpect 4.8.0 pypi_0 pypi
pickleshare 0.7.5 pypi_0 pypi
pillow 10.0.0 pypi_0 pypi
pip 23.1.2 py38h06a4308_0
pkgutil-resolve-name 1.3.10 pypi_0 pypi
platformdirs 3.8.1 pypi_0 pypi
prometheus-client 0.17.1 pypi_0 pypi
prompt-toolkit 3.0.39 pypi_0 pypi
protobuf 4.23.4 pypi_0 pypi
psutil 5.9.5 pypi_0 pypi
ptyprocess 0.7.0 pypi_0 pypi
pure-eval 0.2.2 pypi_0 pypi
pyasn1 0.5.0 pypi_0 pypi
pyasn1-modules 0.3.0 pypi_0 pypi
pyclipper 1.3.0.post4 pypi_0 pypi
pycparser 2.21 pypi_0 pypi
pygments 2.15.1 pypi_0 pypi
pyparsing 3.0.9 pypi_0 pypi
pytesseract 0.3.10 pypi_0 pypi
python 3.8.17 h955ad1f_0
python-bidi 0.4.2 pypi_0 pypi
python-dateutil 2.8.2 pypi_0 pypi
python-json-logger 2.0.7 pypi_0 pypi
pytz 2023.3 pypi_0 pypi
pywavelets 1.4.1 pypi_0 pypi
pyyaml 6.0 pypi_0 pypi
pyzmq 25.1.0 pypi_0 pypi
readline 8.2 h5eee18b_0
referencing 0.29.1 pypi_0 pypi
requests 2.31.0 pypi_0 pypi
requests-oauthlib 1.3.1 pypi_0 pypi
rfc3339-validator 0.1.4 pypi_0 pypi
rfc3986-validator 0.1.1 pypi_0 pypi
rpds-py 0.8.10 pypi_0 pypi
rsa 4.9 pypi_0 pypi
scikit-image 0.21.0 pypi_0 pypi
scipy 1.10.1 pypi_0 pypi
send2trash 1.8.2 pypi_0 pypi
setuptools 67.8.0 py38h06a4308_0
shapely 2.0.1 pypi_0 pypi
six 1.16.0 pypi_0 pypi
sniffio 1.3.0 pypi_0 pypi
soupsieve 2.4.1 pypi_0 pypi
sqlite 3.41.2 h5eee18b_0
stack-data 0.6.2 pypi_0 pypi
sympy 1.12 pypi_0 pypi
tensorboard 2.13.0 pypi_0 pypi
tensorboard-data-server 0.7.1 pypi_0 pypi
tensorflow 2.13.0 pypi_0 pypi
tensorflow-estimator 2.13.0 pypi_0 pypi
tensorflow-io-gcs-filesystem 0.32.0 pypi_0 pypi
termcolor 2.3.0 pypi_0 pypi
terminado 0.17.1 pypi_0 pypi
tifffile 2023.7.10 pypi_0 pypi
tinycss2 1.2.1 pypi_0 pypi
tk 8.6.12 h1ccaba5_0
tomli 2.0.1 pypi_0 pypi
torch 2.0.1 pypi_0 pypi
torchvision 0.15.2 pypi_0 pypi
tornado 6.3.2 pypi_0 pypi
tqdm 4.65.0 pypi_0 pypi
traitlets 5.9.0 pypi_0 pypi
triton 2.0.0 pypi_0 pypi
typing-extensions 4.5.0 pypi_0 pypi
tzdata 2023.3 pypi_0 pypi
uri-template 1.3.0 pypi_0 pypi
urllib3 1.26.16 pypi_0 pypi
validators 0.20.0 pypi_0 pypi
wcwidth 0.2.6 pypi_0 pypi
webcolors 1.13 pypi_0 pypi
webencodings 0.5.1 pypi_0 pypi
websocket-client 1.6.1 pypi_0 pypi
werkzeug 2.3.6 pypi_0 pypi
wheel 0.38.4 py38h06a4308_0
wrapt 1.15.0 pypi_0 pypi
xz 5.4.2 h5eee18b_0
zipp 3.16.2 pypi_0 pypi
zlib 1.2.13 h5eee18b_0
```
|
open
|
2023-07-15T07:11:40Z
|
2023-07-16T05:51:58Z
|
https://github.com/JaidedAI/EasyOCR/issues/1083
|
[] |
nyck33
| 1
|
junyanz/pytorch-CycleGAN-and-pix2pix
|
pytorch
| 1,176
|
l2 regularisation
|
Hello,
I want to add l2 regularisation .Can you tell me where can I add this line:
optimizer = torch.optim.Adam(model.parameters(), lr=1e-4, weight_decay=1e-5)
|
open
|
2020-11-06T20:03:48Z
|
2020-11-25T18:01:54Z
|
https://github.com/junyanz/pytorch-CycleGAN-and-pix2pix/issues/1176
|
[] |
SurbhiKhushu
| 1
|
dropbox/sqlalchemy-stubs
|
sqlalchemy
| 216
|
(Clarification) What LICENSE does this package use?
|
Hi! I'm currently packaging this for [Guix](https://guix.gnu.org/); and the setup file indicates that the project is MIT licensed, yet the actual LICENSE file is Apache 2.0. Could someone please clarify this. Guix review [here](https://patches.guix-patches.cbaines.net/project/guix-patches/patch/20210508204124.38500-2-me@bonfacemunyoki.com/)
|
closed
|
2021-05-10T09:36:10Z
|
2021-05-10T19:47:27Z
|
https://github.com/dropbox/sqlalchemy-stubs/issues/216
|
[] |
BonfaceKilz
| 2
|
WZMIAOMIAO/deep-learning-for-image-processing
|
pytorch
| 807
|
请教vit模型,百度网盘里面的权重是怎么得到的?自己重新训练的还是从官方实现的npz权重转换过来的?
|
def vit_base_patch16_224(num_classes: int = 1000):
"""
ViT-Base model (ViT-B/16) from original paper (https://arxiv.org/abs/2010.11929).
ImageNet-1k weights @ 224x224, source https://github.com/google-research/vision_transformer.
weights ported from official Google JAX impl:
链接: https://pan.baidu.com/s/1zqb08naP0RPqqfSXfkB2EA 密码: eu9f
"""
model = VisionTransformer(img_size=224,
patch_size=16,
embed_dim=768,
depth=12,
num_heads=12,
representation_size=None,
num_classes=num_classes)
return model
百度网盘里的权重,weights ported from official Google JAX impl是什么意思?直接从npz模型转过来的,还是自己重新训练得到了这个模型?
|
closed
|
2024-05-14T07:02:06Z
|
2024-06-26T15:28:49Z
|
https://github.com/WZMIAOMIAO/deep-learning-for-image-processing/issues/807
|
[] |
ShihuaiXu
| 1
|
Asabeneh/30-Days-Of-Python
|
matplotlib
| 526
|
Դասեր
|
closed
|
2024-06-07T07:31:12Z
|
2024-06-07T07:50:12Z
|
https://github.com/Asabeneh/30-Days-Of-Python/issues/526
|
[] |
Goodmood55
| 0
|
|
polarsource/polar
|
fastapi
| 4,744
|
Upcoming deprecation in Pydantic 2.11
|
### Description
We recently added polar in our list of our tested third party libraries, to better prevent regressions in future versions of Pydantic.
To improve build performance, we are going to make some internal changes to the handling of `__get_pydantic_core_schema__` and Pydantic models in https://github.com/pydantic/pydantic/pull/10863. As a consequence, the `__get_pydantic_core_schema__` method of the `BaseModel` class was going to be removed, but turns out that some projects (including polar) are relying on this method, e.g. in the `ListResource` model:
https://github.com/polarsource/polar/blob/ae2c70aeb877969bb2267271cd33cced636e4a2d/server/polar/kit/pagination.py#L146-L155
As a consequence, we are going to raise a deprecation warning when `super().__get_pydantic_core_schema__` is being called to ease transition. In the future, this can be fixed by directly calling `handler(source)` instead. However, I wouldn't recommend implementing `__get_pydantic_core_schema__` on Pydantic models, as it can lead to unexpected behavior.
In the case of `ListResource`, you are mutating the core schema reference, which is crashing the core schema generation logic in some cases:
```python
class ListResource[T](BaseModel):
@classmethod
def __get_pydantic_core_schema__(
cls, source: type[BaseModel], handler: GetCoreSchemaHandler, /
) -> CoreSchema:
"""
Override the schema to set the `ref` field to the overridden class name.
"""
result = super().__get_pydantic_core_schema__(source, handler)
result["ref"] = cls.__name__ # type: ignore
return result
class Model(BaseModel):
a: ListResource[int]
b: ListResource[int]
# Crash with a KeyError when the schema for `Model` is generated
```
The reason for this is that internally, references are used to avoid generating a core schema twice for the same object (in the case of `Model`, the core schema for `ListResource[int]` is only generated once). To do so, we generate a reference for the object and compare it with the already generated definitions. But because the `"ref"` was dynamically changed, Pydantic is not able to retrieve the already generated schema and this breaks a lot of things.
It seems that changing the ref was made in order to simplify the generated JSON Schema names in https://github.com/polarsource/polar/pull/3833. Instead, I would suggest [using a custom `GenerateJsonSchema` class](https://docs.pydantic.dev/latest/concepts/json_schema/#customizing-the-json-schema-generation-process), and overriding the relevant method (probably `get_defs_ref`). I know it may be more tedious to do so, but altering the core schema ref directly is never going to play well [^1]
---
As a side note, I also see you are using the internal `display_as_type` function:
https://github.com/polarsource/polar/blob/ae2c70aeb877969bb2267271cd33cced636e4a2d/server/polar/kit/pagination.py#L127-L141
Because `ListResource` is defined with a single type variable, I can suggest using the following instead:
```python
@classmethod
def model_parametrized_name(cls, params: tuple[type[Any]]) -> str: # Guaranteed to be of length 1
"""
Override default model name implementation to detect `ClassName` metadata.
It's useful to shorten the name when a long union type is used.
"""
param = params[0]
if hasattr(param, "__metadata__"):
for metadata in param.__metadata__:
if isinstance(metadata, ClassName):
return f"{cls.__name__}[{metadata.name}]"
return super().model_parametrized_name(params)
```
But, again, if this is done for JSON Schema generation purposes, it might be best to leave the model name unchanged and define a custom `GenerateJsonSchema` class.
[^1]: Alternatively, we are thinking about designing a new API for core schema generation, that would allow providing a custom reference generation implementation for Pydantic models (but also other types).
|
open
|
2025-01-01T19:52:49Z
|
2025-01-10T21:22:58Z
|
https://github.com/polarsource/polar/issues/4744
|
[
"bug"
] |
Viicos
| 2
|
tflearn/tflearn
|
tensorflow
| 896
|
Decoder output giving wrong result in textsum
|
Hi,michaelisard
I trained the model using toy dataset .when I am trying to test using one article in decode ,then decoder is giving the other article as output which is not in test data .when i tried it with 5000 epochs
Eg: abstract=<d> <p> <s> sri lanka closes schools as war escalates . </s> </p> </d> article=<d> <p> <s> the sri lankan government on wednesday announced the closure of government schools with immediate effect as a military campaign against tamil separatists escalated in the north of the country . </s> <s> the cabinet wednesday decided to advance the december holidays by one month because of a threat from the liberation tigers of tamil eelam -lrb- ltte -rrb- against school children , a government official said . </s> <s> `` there are intelligence reports that the tigers may try to kill a lot of children to provoke a backlash against tamils in colombo . </s> <s> `` if that happens , troops will have to be withdrawn from the north to maintain law and order here , '' a police official said . </s> <s> he said education minister richard pathirana visited several government schools wednesday before the closure decision was taken . </s> <s> the government will make alternate arrangements to hold end of term examinations , officials said . </s> <s> earlier wednesday , president chandrika kumaratunga said the ltte may step up their attacks in the capital to seek revenge for the ongoing military offensive which she described as the biggest ever drive to take the tiger town of jaffna . . </s> </p> </d> publisher=AFP
output:output=financial markets end turbulent week equipment business .
|
open
|
2017-09-02T11:36:24Z
|
2017-09-02T11:36:24Z
|
https://github.com/tflearn/tflearn/issues/896
|
[] |
ashakodali
| 0
|
pytest-dev/pytest-html
|
pytest
| 476
|
Lines on both \r and \n
|
html report for log add lines for both \r and \n. This seems to be a bug.
Suppose we had a test:
def test_01_print():
print("one")
print("two\r\n", end="")
print("three\raaa")
print("four")
assert True
Terminal would show:
__________test_01_print ________
----- Captured stdout call -----
one
two
aaaee
four
However, report adds a line with \r
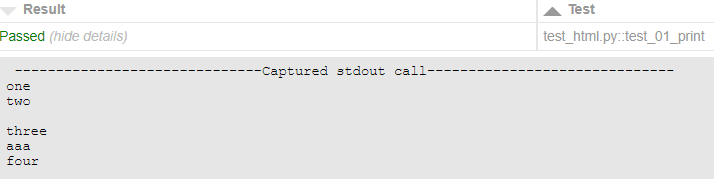
|
open
|
2021-12-28T06:37:43Z
|
2021-12-28T08:00:24Z
|
https://github.com/pytest-dev/pytest-html/issues/476
|
[] |
listerplus
| 1
|
CorentinJ/Real-Time-Voice-Cloning
|
deep-learning
| 577
|
List python/whatever versions required for this to work
|
Could you list proper versions of the required components?
There seem to be a hundred versions of Python, not to mention there's something called "Python 2" and "Python 3". Even minor versions of these thingies have their own incompatibilities and quirks.
I keep getting there's no package for TensorFlow for my config, I've tried a lot of combinations with x64 Pythons for about an hour. Now I'm giving up, it's hopeless...
|
closed
|
2020-10-28T09:56:55Z
|
2020-10-28T15:21:07Z
|
https://github.com/CorentinJ/Real-Time-Voice-Cloning/issues/577
|
[] |
ssuukk
| 2
|
JaidedAI/EasyOCR
|
pytorch
| 530
|
Question regarding training custom model
|
To the concerned,
* Just took a look at your training custom model section
* Reviewed the sample dataset and corresponding pth file. I see in your data set each jpg contained is one single word. I am not sure how to go about generating such a dataset in my case from P&ID engineering diagrams (printed not hand drawn/written).
* What do you suggest in my case?
* How to go about?
* Currently
-- some of the vertical text in the diagram is not getting detected - this just using whatever default detector and recognizer models that EasyOCR downloads on readtext.
-- also there are overlapping detections, for e.g. I have a valve part that has the tag 14-HV-334-02, sometimes the detections are 14-, HV-334 (with -02 not all detected) or may be with overlap as 14-HV and HV-334-02 or some combination of...
Please advise. I can post screen shots, if you need.
|
closed
|
2021-08-30T15:27:44Z
|
2021-10-06T08:52:41Z
|
https://github.com/JaidedAI/EasyOCR/issues/530
|
[] |
pankaja0285
| 2
|
GibbsConsulting/django-plotly-dash
|
plotly
| 464
|
Having issues with dynamically loading images in dash app. Demo-nine missing
|
Trying to port existing dash apps into dpd. I'm having an issue with displaying images or other media in my django dash app. The documentation doesn't say much on the matter. The dash app requests are return 404 not found errors even though the files are in the assets folders. Have also tried with static folders. I also noticed that demo-nine's images are coming up as broken links and demo-nine is commented out from the live demo. My use case is for the user to upload and view files through the dash app. Is this supported or am I using django-plotly-dash for the wrong use case.
|
closed
|
2023-06-12T20:10:12Z
|
2024-10-01T03:33:00Z
|
https://github.com/GibbsConsulting/django-plotly-dash/issues/464
|
[] |
giaxle
| 2
|
onnx/onnx
|
scikit-learn
| 6,290
|
Inference session with 'optimized_model_filepath' expands Gelu operator into atomic operators
|
# Inference session with `optimized_model_filepath` expands Gelu operator into atomic operators. Can we turn this off?
### Question
When I create InferenceSession and specify `optimized_model_filepath` in `SessionOptions`, Gelu operator is expanded into atomic operators. Same happens also for HardSwish. Can this optimization be turned off, so the optimized model contains only single Gelu operator?
I went through available flags in `SessionOptions` but nothing seems to resolve this. I also tried to disable all optimizations via attribute `disabled_optimizers` but this doesn't work either.
### Further information
Code to reproduce:
```python
import onnx
import onnxruntime
from onnx import TensorProto
graph = onnx.helper.make_graph(
[onnx.helper.make_node("Gelu", ["x"], ["output"])],
'Gelu test',
[onnx.helper.make_tensor_value_info("x", TensorProto.FLOAT, [10])],
[onnx.helper.make_tensor_value_info("output", TensorProto.FLOAT, ())],
)
onnx_model = onnx.helper.make_model(graph)
sess_options = onnxruntime.SessionOptions()
sess_options.optimized_model_filepath = "expanded_gelu.onnx"
sess_options.graph_optimization_level = onnxruntime.GraphOptimizationLevel.ORT_DISABLE_ALL
optimizers = [
"AttentionFusion", "Avx2WeightS8ToU8Transformer", "BiasDropoutFusion", "BiasGeluFusion", "BiasSoftmaxDropoutFusion",
"BiasSoftmaxFusion", "BitmaskDropoutReplacement", "CommonSubexpressionElimination", "ConcatSliceElimination",
"ConstantFolding", "ConstantSharing", "Conv1dReplacement", "DoubleQDQPairsRemover", "DummyGraphTransformer",
"DynamicQuantizeMatMulFusion", "EmbedLayerNormFusion", "EnsureUniqueDQForNodeUnit", "FastGeluFusion",
"FreeDimensionOverrideTransformer", "GatherToSplitFusion", "GatherToSliceFusion", "GeluApproximation", "GeluFusion",
"GemmActivationFusion", "TrainingGraphTransformerConfiguration", "IdenticalChildrenConsolidation",
"RemoveDuplicateCastTransformer", "IsInfReduceSumFusion", "LayerNormFusion", "SimplifiedLayerNormFusion",
"GeluRecompute", "AttentionDropoutRecompute", "SoftmaxCrossEntropyLossInternalFusion", "MatMulActivationFusion",
"MatMulAddFusion", "MatMulIntegerToFloatFusion", "MatMulScaleFusion", "MatmulTransposeFusion",
"MegatronTransformer", "MemoryOptimizer", "NchwcTransformer", "NhwcTransformer", "PaddingElimination",
"PropagateCastOps", "QDQFinalCleanupTransformer", "QDQFusion", "QDQPropagationTransformer", "QDQS8ToU8Transformer",
"QuickGeluFusion", "ReshapeFusion", "RocmBlasAltImpl", "RuleBasedGraphTransformer", "ScaledSumFusion",
"SceLossGradBiasFusion", "InsertGatherBeforeSceLoss", "SelectorActionTransformer", "ShapeOptimizer",
"SkipLayerNormFusion", "TransformerLayerRecompute", "MemcpyTransformer", "TransposeOptimizer", "TritonFusion",
"UpStreamGraphTransformerBase"]
sess = onnxruntime.InferenceSession(onnx_model.SerializeToString(), sess_options, disabled_optimizers=optimizers)
```
`expanded_gelu.onnx`:

### Notes
onnx=1.15.0
onnxruntime=1.17.3
|
closed
|
2024-08-09T11:33:44Z
|
2024-08-12T06:58:27Z
|
https://github.com/onnx/onnx/issues/6290
|
[
"question"
] |
skywall
| 0
|
lux-org/lux
|
jupyter
| 159
|
ERROR:root:Internal Python error in the inspect module.
|
Printing out Vis and VisList occasionally results in this error. It is unclear where this traceback error is coming from.


|
closed
|
2020-11-28T09:44:17Z
|
2020-11-28T12:05:35Z
|
https://github.com/lux-org/lux/issues/159
|
[
"bug",
"priority"
] |
dorisjlee
| 2
|
jupyterhub/jupyterhub-deploy-docker
|
jupyter
| 69
|
Fails if behind proxy
|
This currently doesn't work if you're on a proxied network.
I've made some changes that have fixed most of it, but I haven't completely gotten it working yet.
This is from the `make notebook_image` command.
```
e1677043235c: Pulling from jupyter/minimal-notebook
Status: Image is up to date for jupyter/minimal-notebook:e1677043235c
docker build -t jupyterhub-user \
--build-arg JUPYTERHUB_VERSION=0.8.0 \
--build-arg DOCKER_NOTEBOOK_IMAGE=jupyter/minimal-notebook:e1677043235c \
singleuser
Sending build context to Docker daemon 2.048kB
Step 1/4 : ARG DOCKER_NOTEBOOK_IMAGE
Step 2/4 : FROM $DOCKER_NOTEBOOK_IMAGE
---> c86d7e5f432a
Step 3/4 : ARG JUPYTERHUB_VERSION
---> Using cache
---> be71e0724cd5
Step 4/4 : RUN python3 -m pip install --no-cache jupyterhub==$JUPYTERHUB_VERSION
---> Running in 02c9c26fcc9b
Collecting jupyterhub==0.8.0
Retrying (Retry(total=4, connect=None, read=None, redirect=None)) after connection broken by 'ConnectTimeoutError(<pip._vendor.requests.packages.urllib3.connection.VerifiedHTTPSConnection object at 0x7f2f0f764710>, 'Connection to pypi.python.org timed out. (connect timeout=15)')': /simple/jupyterhub/
```
|
closed
|
2018-04-03T07:41:34Z
|
2018-04-04T04:17:09Z
|
https://github.com/jupyterhub/jupyterhub-deploy-docker/issues/69
|
[] |
moppymopperson
| 3
|
grillazz/fastapi-sqlalchemy-asyncpg
|
pydantic
| 174
|
implement scheduler
|
https://apscheduler.readthedocs.io/en/3.x/
|
closed
|
2024-10-09T09:58:09Z
|
2024-10-16T13:37:36Z
|
https://github.com/grillazz/fastapi-sqlalchemy-asyncpg/issues/174
|
[] |
grillazz
| 0
|
dropbox/PyHive
|
sqlalchemy
| 120
|
Extra requirements are not installed on Python 3.6
|
I'm trying to install pyhive with hive interfaces with pip.
But pip does not install extra requirements for hive interface.
Here's my shell output with warning messages
```
(venv) $ python --version
Python 3.6.1
(venv) $ pip --version
pip 9.0.1 from /Users/owen/.virtualenvs/venv/lib/python3.6/site-packages (python 3.6)
(venv) $ pip install pyhive[hive]
Requirement already satisfied: pyhive[hive] in /Users/owen/.virtualenvs/venv/lib/python3.6/site-packages
Ignoring sasl: markers 'extra == "Hive"' don't match your environment
Ignoring thrift-sasl: markers 'extra == "Hive"' don't match your environment
Ignoring thrift: markers 'extra == "Hive"' don't match your environment
Requirement already satisfied: future in /Users/owen/.virtualenvs/venv/lib/python3.6/site-packages (from pyhive[hive])
```
I'm working on macOS 10.12.4 and Python 3.6.1 installed by pyenv.
`sasl`, `thrift-sasl`, `thrift` are installed very well when I try to install them manually (`pip install sasl ...`)
It works very well when I try to install in Python 2.7.11 installed by pyenv.
|
closed
|
2017-05-16T04:31:19Z
|
2017-05-16T23:51:16Z
|
https://github.com/dropbox/PyHive/issues/120
|
[] |
Hardtack
| 2
|
gradio-app/gradio
|
python
| 10,267
|
gradio 5.0 unable to load javascript file
|
### Describe the bug
if I provide JavaScript code in a variable, it is executed perfectly well but when I put the same code in a file "app.js" and then pass the file path in `js` parameter in `Blocks`, it doesn't work. I have added the code in reproduction below. if the same code is put in a file, the block will be unable to execute that.
It was working fine in version 4. Now I am upgrading to 5.0.
### Have you searched existing issues? 🔎
- [X] I have searched and found no existing issues
### Reproduction
```python
import gradio as gr
login_page_js = """
() => {
//handle launch
let reload = false;
let gradioURL = new URL(window.location.href);
if(
!gradioURL.searchParams.has('__theme') ||
(gradioURL.searchParams.has('__theme') && gradioURL.searchParams.get('__theme') !== 'dark')
) {
gradioURL.searchParams.delete('__theme');
gradioURL.searchParams.set('__theme', 'dark');
reload = true;
}
if(reload) {
window.location.replace(gradioURL.href);
}
}
"""
with gr.Blocks(
js = login_page_js
) as login_page:
gr.Button("Sign in with Microsoft", elem_classes="icon-button" ,link="/login")
if __name__ == "__main__":
login_page.launch()
```
### Screenshot
_No response_
### Logs
_No response_
### System Info
```shell
linux 2204
```
### Severity
I can work around it
|
open
|
2024-12-30T15:09:28Z
|
2024-12-30T16:19:48Z
|
https://github.com/gradio-app/gradio/issues/10267
|
[
"bug"
] |
git-hamza
| 2
|
alirezamika/autoscraper
|
web-scraping
| 35
|
ERROR: Package 'autoscraper' requires a different Python: 2.7.16 not in '>=3.6'
|
All 3 listed installation methods return the error shown in the issue title & cause an installation failure. No change when using pip or pip3 command.
I tried running the following 2 commands to get around the pre-commit issue but with no change in the result:
$ pip uninstall pre-commit # uninstall from Python2.7
$ pip3 install pre-commit # install with Python3
|
closed
|
2020-10-28T23:40:46Z
|
2020-10-29T14:58:19Z
|
https://github.com/alirezamika/autoscraper/issues/35
|
[] |
mechengineermike
| 4
|
NullArray/AutoSploit
|
automation
| 703
|
Divided by zero exception23
|
Error: Attempted to divide by zero.23
|
closed
|
2019-04-19T15:59:23Z
|
2019-04-19T16:38:55Z
|
https://github.com/NullArray/AutoSploit/issues/703
|
[] |
AutosploitReporter
| 0
|
skypilot-org/skypilot
|
data-science
| 4,728
|
Restart an INIT cluster skips file_mounts and setup
|
## Actual Behavior
Launched a cluster but failed when syncing file mounts:
```
$ sky launch -c aylei-cs api-server-test.yaml
YAML to run: api-server-test.yaml
Considered resources (1 node):
---------------------------------------------------------------------------------------------
CLOUD INSTANCE vCPUs Mem(GB) ACCELERATORS REGION/ZONE COST ($) CHOSEN
---------------------------------------------------------------------------------------------
AWS c6i.2xlarge 8 16 - ap-southeast-1 0.39 ✔
---------------------------------------------------------------------------------------------
Launching a new cluster 'aylei-cs'. Proceed? [Y/n]: Y
⚙︎ Launching on AWS ap-southeast-1 (ap-southeast-1a,ap-southeast-1b,ap-southeast-1c).
├── Instance is up.
└── Docker container is up.
Open file descriptor limit (256) is low. File sync to remote clusters may be slow. Consider increasing the limit using `ulimit -n <number>` or modifying system limits.
✓ Cluster launched: aylei-cs. View logs: sky api logs -l sky-2025-02-16-22-16-56-301393/provision.log
⚙︎ Syncing files.
Syncing (to 1 node): /Users/aylei/repo/skypilot-org/skypilot -> ~/.sky/file_mounts/sky_repo
sky.exceptions.CommandError: Command rsync -Pavz --filter='dir-merge,- .gitignore' -e 'ssh -i /Users/aylei/.sky/clients/57339c81/ssh/sky-... failed with return code 255.
Failed to rsync up: /Users/aylei/repo/skypilot-org/skypilot -> ~/.sky/file_mounts/sky_repo. Ensure that the network is stable, then retry.
```
Restarted the cluster in `INIT` state:
```
sky start aylei-cs
Restarting 1 cluster: aylei-cs. Proceed? [Y/n]: Y
⚙︎ Launching on AWS ap-southeast-1 (ap-southeast-1a).
├── Instance is up.
└── Docker container is up.
Open file descriptor limit (256) is low. File sync to remote clusters may be slow. Consider increasing the limit using `ulimit -n <number>` or modifying system limits.
✓ Cluster launched: aylei-cs. View logs: sky api logs -l sky-2025-02-16-22-23-19-710647/provision.log
Cluster aylei-cs started.
```
SSH to the cluster, observed no `/sky_repo` mounted and no setup executed.
```
ssh aylei-cs
ls /sky_repo
ls: cannot access '/sky_repo': No such file or directory
```
## Expected Behavior
I expect skypilot either:
- setup `file_mounts` and run setup job idempotently on cluster restart
- or warn user that the state might be broken when restarting a cluster in `INIT` state and hint user re-launch the cluster (`sky down && sky launch`) to get a clean state
## Appendix
The sky yaml I used:
```yaml
resources:
cloud: aws
cpus: 8+
memory: 16+
ports: 46580
image_id: docker:berkeleyskypilot/skypilot-beta:latest
region: ap-southeast-1
file_mounts:
/sky_repo: /Users/aylei/repo/skypilot-org/skypilot
~/.ssh/id_rsa.pub: ~/.ssh/id_rsa.pub
envs:
SKYPILOT_POD_CPU_CORE_LIMIT: "7"
SKYPILOT_POD_MEMORY_GB_LIMIT: "14"
setup: |
cd /sky_repo
pip install -r requirements-dev.txt
pip install -e .[aws]
mkdir -p ~/.sky
# Add any custom config here
cat <<EOF > ~/.sky/config.yaml
allowed_clouds:
- gcp
- aws
- kubernetes
EOF
sky api start --deploy
```
|
open
|
2025-02-16T15:53:21Z
|
2025-02-20T05:11:01Z
|
https://github.com/skypilot-org/skypilot/issues/4728
|
[
"bug",
"core"
] |
aylei
| 2
|
marshmallow-code/flask-smorest
|
rest-api
| 55
|
Fix base path when script root present
|
Same as flask_apispec's issue: https://github.com/jmcarp/flask-apispec/pull/125/files
The application may be deployed under a path, such as when deploying as a serverless lambda. The path needs to be prefixed.
|
closed
|
2019-04-03T08:21:47Z
|
2021-04-29T07:53:15Z
|
https://github.com/marshmallow-code/flask-smorest/issues/55
|
[] |
revmischa
| 25
|
litestar-org/litestar
|
pydantic
| 3,764
|
Enhancement: local state for websocket listeners
|
### Summary
There seems to be no way to have a state that is unique to a particular websocket connection. Or maybe it's possible, but it's not documented?
### Basic Example
Consider the following example:
```
class Listener(WebsocketListener):
path = "/ws"
def on_accept(self, socket: WebSocket, state: State):
state.user_id = str(uuid.uuid4())
def on_receive(self, data: str, state: State):
logger.info("Received: %s", data)
return state.user_id
```
Here, I expect a different `user_id` for each connection, but it turns out to not be the case:
```
def test_listener(app: Litestar):
client_1 = TestClient(app)
client_2 = TestClient(app)
with (
client_1.websocket_connect("/ws") as ws_1,
client_2.websocket_connect("/ws") as ws_2,
):
ws_1.send_text("Hello")
ws_2.send_text("Hello")
assert ws_1.receive_text() != ws_2.receive_text() # FAILS
```
I figured that there is a way to define a custom `Websocket` class which seems to be bound to a particular connection, but if that's the only way, should it be that hard for such a common use case?
UPDATE:
`WebSocketScope` contains it's own state dict, and it's unique to each connection. So my suggestion is to resolve the `state` dependency corresponding to the scope in that case.
|
closed
|
2024-09-28T15:00:48Z
|
2025-03-20T15:54:56Z
|
https://github.com/litestar-org/litestar/issues/3764
|
[
"Enhancement"
] |
olzhasar
| 4
|
adamerose/PandasGUI
|
pandas
| 222
|
Having following issue on install of pandasGUI
|
### Discussed in https://github.com/adamerose/PandasGUI/discussions/221
<div type='discussions-op-text'>
<sup>Originally posted by **decisionstats** March 6, 2023</sup>
ModuleNotFoundError: No module named 'PyQt5.sip'
Uninstalling and reinstalling PyQt did not work for me . Please help in how to install on Windows</div>
|
open
|
2023-03-06T14:25:01Z
|
2023-03-06T14:25:01Z
|
https://github.com/adamerose/PandasGUI/issues/222
|
[] |
decisionstats
| 0
|
nvbn/thefuck
|
python
| 1,349
|
make appimage or binary file
|
appimage format can run everywhere
so , we can not use apt-get install, just download one file
|
open
|
2022-12-11T04:25:47Z
|
2022-12-11T04:25:47Z
|
https://github.com/nvbn/thefuck/issues/1349
|
[] |
newyorkthink
| 0
|
AirtestProject/Airtest
|
automation
| 828
|
1.1.6版本basetouch无效求助
|
升级到1.1.6版本后,使用如下代码模拟拖动图标操作
longtouch_event = [
DownEvent([908, 892]),# 待删除应用的坐标
SleepEvent(2),
MoveEvent([165,285]),# 删除应用的垃圾桶坐标
UpEvent(0)]
dev.touch_proxy.perform(longtouch_event)
实际效果为仅拖动了下拉栏的第一行,如下
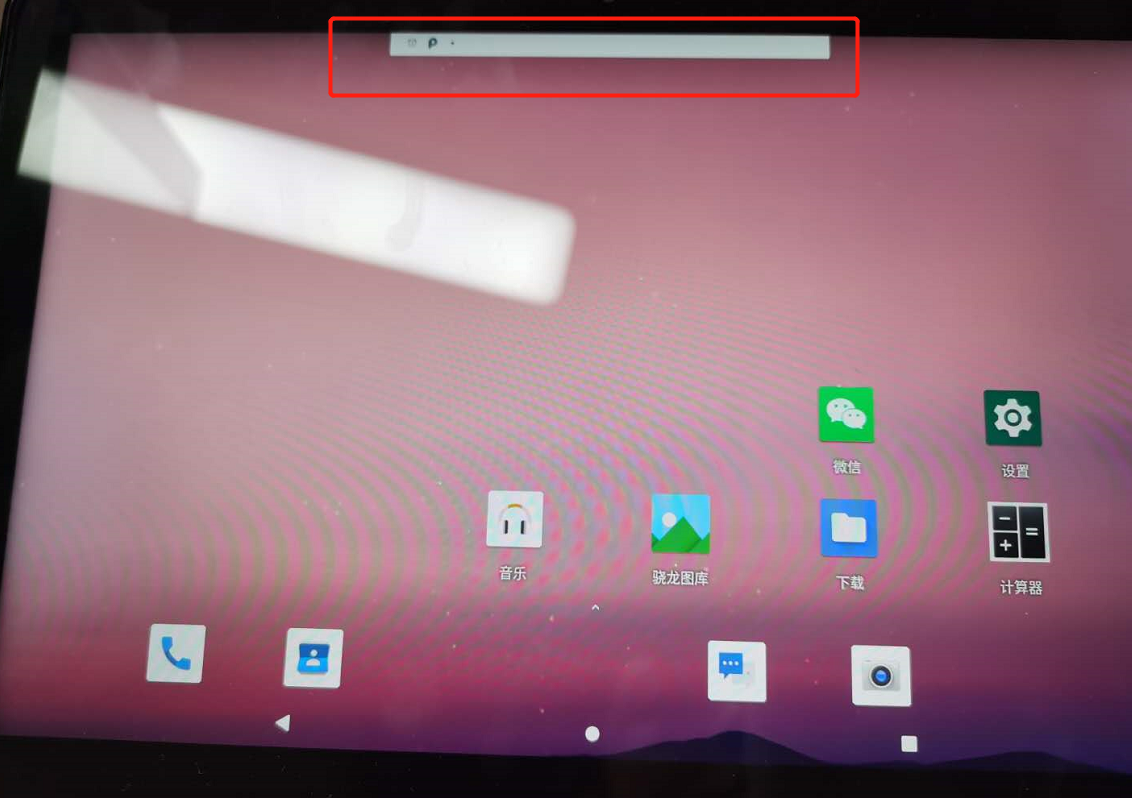
本地报错如下:
Note: device /dev/input/mouse0 is not supported by libevdev'
Note: device /dev/input/mice is not supported by libevdev'
尝试了安卓7/9的多个机器,现象一样
|
closed
|
2020-11-11T07:01:11Z
|
2021-02-21T03:20:15Z
|
https://github.com/AirtestProject/Airtest/issues/828
|
[] |
masterKLB
| 1
|
Lightning-AI/pytorch-lightning
|
data-science
| 19,699
|
Modules with `nn.Parameter` not Converted by Lightning Mixed Precision
|
### Bug description
I have an `nn.Module` (call it `Mod`) which adds its input `x` to an internal `nn.Parameter`. I'm using `Mod` as part of a `pl.LightningModule` which I'm training in `16-mixed` precision. However, the output of calling `Mod` is a tensor with dtype `torch.float32`. When I use other layer types, they output `torch.float16` tensors as expected. This failure is often silent (as in the example provided below), but can cause issues if a model contains a component (e.g. flash attention) that requires fp16. Furthermore, after loading a model trained this way at inference time and calling `.half()` on it, the output is `NaN` or otherwise nonsensical, despite being perfectly fine if I load the model in fp32.
### What version are you seeing the problem on?
v2.0
### How to reproduce the bug
This is a small, reproducible example with `lightning==2.0.2`. Note how the output of `Mod` has dtype `torch.float32` while the output of a linear layer has dtype `torch.float16`. The example runs distributed on 8 GPUs, but the issue is the same on a single GPU.
```python
from lightning import pytorch as pl
import torch
from torch import nn
from torch.utils.data import TensorDataset, DataLoader
class Mod(nn.Module):
def __init__(self):
super().__init__()
derp = torch.randn((1, 32))
self.p = nn.Parameter(derp, requires_grad=False)
def forward(self, x):
return x + self.p
class Model(pl.LightningModule):
def __init__(self):
super().__init__()
self.lin = nn.Linear(32, 32)
self.m = Mod()
self.l = nn.MSELoss()
def forward(self, x):
print('x', x.dtype)
y = self.lin(x)
print('y', y.dtype)
z = self.m(y)
print('z', z.dtype)
print('p',self.m.p.dtype)
print('lin', self.lin.weight.dtype)
return z
def training_step(self, batch, batch_idx):
x, y = batch
z = self(x)
loss = self.l(z, y)
return loss
def configure_optimizers(self):
return torch.optim.Adam(self.parameters())
xdata = torch.randn((1000, 32))
ydata = xdata + torch.randn_like(xdata) * .1
dataset=TensorDataset(xdata,ydata)
dataloader=DataLoader(dataset, batch_size=8, num_workers=4, pin_memory=True)
model = Model()
trainer = pl.Trainer(
strategy='ddp',
accelerator='gpu',
devices=list(range(8)),
precision='16-mixed'
)
trainer.fit(model=model, train_dataloaders=dataloader)
```
### Error messages and logs
Example output:
```
Epoch 3: 78%|███████████████████████████████████████████████████████████████████████████████████████████████████████████████████▏ | 98/125 [00:00<00:00, 138.94it/s, v_num=5]x torch.float32
x torch.float32
y torch.float16
z torch.float32
p torch.float32
lin torch.float32
```
### Environment
<details>
<summary>Current environment</summary>
```
#- Lightning Component (e.g. Trainer, LightningModule, LightningApp, LightningWork, LightningFlow): Trainer, LightningModule
#- PyTorch Lightning Version (e.g., 1.5.0): 2.0.2
#- PyTorch Version (e.g., 2.0): 2.1.0
#- Python version (e.g., 3.9): 3.10.12
#- OS: Ubuntu 20.04.6 LTS (Focal Fossa)
#- CUDA/cuDNN version: 11.8
#- GPU models and configuration: 8xA100
#- How you installed Lightning(`conda`, `pip`, source): pip
```
</details>
### More info
Thank you for your help!
|
open
|
2024-03-25T21:32:26Z
|
2024-03-25T23:24:48Z
|
https://github.com/Lightning-AI/pytorch-lightning/issues/19699
|
[
"question",
"ver: 2.0.x"
] |
nrocketmann
| 2
|
nerfstudio-project/nerfstudio
|
computer-vision
| 2,916
|
Unable to render to mp4 with `RuntimeError: stack expects a non-empty TensorList`
|
**Describe the bug**
Tried to render to MP4. got this error:
```
✅ Done loading checkpoint from outputs/lego_processed/nerfacto/2024-02-14_193442/nerfstudio_models/step-000029999.ckpt
Traceback (most recent call last):
File "/usr/local/bin/ns-render", line 8, in <module>
sys.exit(entrypoint())
File "/usr/local/lib/python3.10/dist-packages/nerfstudio/scripts/render.py", line 896, in entrypoint
tyro.cli(Commands).main()
File "/usr/local/lib/python3.10/dist-packages/nerfstudio/scripts/render.py", line 456, in main
camera_path = get_path_from_json(camera_path)
File "/usr/local/lib/python3.10/dist-packages/nerfstudio/cameras/camera_paths.py", line 178, in get_path_from_json
camera_to_worlds = torch.stack(c2ws, dim=0)
RuntimeError: stack expects a non-empty TensorList
root@3eada9a39237:/workspace# ns-render camera-path --load-config outputs/lego_processed/nerfacto/2024-02-14_193442/config.yml --camera-path-filename /workspace/lego_processed/camera_paths/2024-02-14-19-34-49.json --output-path renders/lego_processed/2024-02-14-19-34-49.mp4
```
**To Reproduce**
1. Download images for training. https://files.extrastatic.dev/extrastatic/Photos-001.zip. Unzip into a directory called lego
1. ` ns-process-data images --data lego/ --output-dir lego_processed`
2. `ns-train nerfacto --data lego_processed/`
3. When completed, try to run render command: `ns-render camera-path --load-config outputs/lego_processed/nerfacto/2024-02-14_193442/config.yml --camera-path-filename /workspace/lego_processed/camera_paths/2024-02-14-19-34-49.json --output-path renders/lego_processed/2024-02-14-19-34-49.mp4`
**Expected behavior**
It should create the mp4 file.
**Additional context**
Using docker:
```
docker build \
--build-arg CUDA_VERSION=11.8.0 \
--build-arg CUDA_ARCHITECTURES=86 \
--build-arg OS_VERSION=22.04 \
--tag nerfstudio-86:0.0.1 \
--file Dockerfile .
```
Inside container `nvidia-smi`:
```
# nvidia-smi
Wed Feb 14 20:39:13 2024
+---------------------------------------------------------------------------------------+
| NVIDIA-SMI 545.29.02 Driver Version: 545.29.02 CUDA Version: 12.3 |
|-----------------------------------------+----------------------+----------------------+
| GPU Name Persistence-M | Bus-Id Disp.A | Volatile Uncorr. ECC |
| Fan Temp Perf Pwr:Usage/Cap | Memory-Usage | GPU-Util Compute M. |
| | | MIG M. |
|=========================================+======================+======================|
| 0 NVIDIA GeForce RTX 3090 Off | 00000000:0A:00.0 Off | N/A |
| 0% 41C P8 23W / 370W | 5MiB / 24576MiB | 0% Default |
| | | N/A |
+-----------------------------------------+----------------------+----------------------+
+---------------------------------------------------------------------------------------+
| Processes: |
| GPU GI CI PID Type Process name GPU Memory |
| ID ID Usage |
|=======================================================================================|
| No running processes found |
+---------------------------------------------------------------------------------------+
```
`colmap -h`
```
# colmap -h
COLMAP 3.8 -- Structure-from-Motion and Multi-View Stereo
(Commit 43de802 on 2023-01-31 with CUDA)
Usage:
colmap [command] [options]
Documentation:
https://colmap.github.io/
Example usage:
colmap help [ -h, --help ]
colmap gui
colmap gui -h [ --help ]
colmap automatic_reconstructor -h [ --help ]
colmap automatic_reconstructor --image_path IMAGES --workspace_path WORKSPACE
colmap feature_extractor --image_path IMAGES --database_path DATABASE
colmap exhaustive_matcher --database_path DATABASE
colmap mapper --image_path IMAGES --database_path DATABASE --output_path MODEL
...
Available commands:
help
gui
automatic_reconstructor
bundle_adjuster
color_extractor
database_cleaner
database_creator
database_merger
delaunay_mesher
exhaustive_matcher
feature_extractor
feature_importer
hierarchical_mapper
image_deleter
image_filterer
image_rectifier
image_registrator
image_undistorter
image_undistorter_standalone
mapper
matches_importer
model_aligner
model_analyzer
model_comparer
model_converter
model_cropper
model_merger
model_orientation_aligner
model_splitter
model_transformer
patch_match_stereo
point_filtering
point_triangulator
poisson_mesher
project_generator
rig_bundle_adjuster
sequential_matcher
spatial_matcher
stereo_fusion
transitive_matcher
vocab_tree_builder
vocab_tree_matcher
vocab_tree_retriever
```
Last commit from `git log:`
```
commit 4f798b23f6c65ef2970145901a0251b61ec8a447 (HEAD -> main, origin/main, origin/HEAD)
Author: Sebastiaan <751205+SharkWipf@users.noreply.github.com>
Date: Tue Feb 13 19:09:59 2024 +0100
...
```
|
open
|
2024-02-14T20:43:12Z
|
2024-05-09T13:32:28Z
|
https://github.com/nerfstudio-project/nerfstudio/issues/2916
|
[] |
xrd
| 5
|
pallets/flask
|
flask
| 5,119
|
Flask 2.3.2 is not compatible with Flassger
|
Flask 2.3.2 is not compatible with Flassger
Description:
After upgrading Flask to latest version we are getting below error.
```
from flasgger import Swagger
File "C:\Python310\lib\site-packages\flasgger\__init__.py", line 10, in <module>
from .base import Swagger, Flasgger, NO_SANITIZER, BR_SANITIZER, MK_SANITIZER, LazyJSONEncoder # noqa
File "C:\Python310\lib\site-packages\flasgger\base.py", line 28, in <module>
from flask.json import JSONEncoder
ImportError: cannot import name 'JSONEncoder' from 'flask.json' (C:\Python310\lib\site-packages\flask\json\__init__.py)
```
After downgrading flask to 2.2.3, it works again!
Environment:
- Python version: 3.10.10
- Flask version: 2.3.2
- Flassger: Latest
|
closed
|
2023-05-09T10:04:25Z
|
2023-05-24T00:05:28Z
|
https://github.com/pallets/flask/issues/5119
|
[] |
nagamanickamm
| 1
|
Josh-XT/AGiXT
|
automation
| 1,367
|
`Providers` has Nested GQL Key
|
### Description
`providers` is the only object that has itself as a subkey in GQL.

### Operating System
- [x] Linux
- [ ] Windows
- [ ] MacOS
### Acknowledgements
- [x] I am NOT trying to use localhost for providers running outside of the docker container.
- [x] I am NOT trying to run AGiXT outside of docker, the only supported method to run it.
- [x] Python 3.10.X is installed and the version in use on the host machine.
- [x] The latest version of Docker is installed and running on the host machine.
- [x] I am using the latest stable version of AGiXT.
- [x] I have provided enough information for the maintainers to reproduce and diagnose the issue.
- [x] I have searched the existing issues to make sure this bug has not been reported yet.
|
closed
|
2025-01-17T22:42:55Z
|
2025-01-19T00:46:00Z
|
https://github.com/Josh-XT/AGiXT/issues/1367
|
[
"type | report | bug",
"area | back end"
] |
JamesonRGrieve
| 0
|
christabor/flask_jsondash
|
plotly
| 83
|
Add Vega support
|
https://vega.github.io/vega-lite
|
closed
|
2017-02-11T01:10:22Z
|
2017-05-03T06:47:31Z
|
https://github.com/christabor/flask_jsondash/issues/83
|
[] |
christabor
| 2
|
pallets-eco/flask-sqlalchemy
|
flask
| 1,289
|
Potential memory leak with table polymorphism
|
## Description
<!--Describe the bug clearly and concisely. Include screenshots if possible-->
I have a Flask application interacting with a PostgreSQL database, and I'm using Flask-SQLAlchemy to define my schemas and queries. I have multiple tables and a polymorphic relationship between them, and what I've realised is that when I call a Flask route that queries large amounts of rows on a table that's related to other with some polymorphic relationship, the RAM allocated to the query does not get released properly even after the request has completed and even if I try to manually call the garbage collector or delete the variable that was holding the query response.
I have tried narrowing down the problem to the simplest chunk of code that still produces the memory leak, and I observe that when I either remove the polymorphic relationship between my tables or simply use SQLAlchemy to make the same computations, the memory is released correctly and goes back down to its pre-query consumption, which makes me believe it is related to the Flask-SQLAlchemy library and polymorphy.
## Reproduce
<!--Describe step-by-step instructions to reproduce the behavior-->
As the problem is not super trivial to reproduce, I added the code for both scenarios :
- Only with SQLAlchemy : <a href='https://github.com/Rmarieta/sqlalchemy-leak'>Repository link</a>.
- With Flask-SQLAlchemy : <a href='https://github.com/Rmarieta/flask-sqlalchemy-leak'>Repository link</a>.
1. In both cases, I build two containers, one for the database and one for either the python process that will use SQLAlchemy or the Flask process using Flask-SQLAlchemy.
2. The containers can be started with `docker-compose up --build`
3. It automatically populates the Event table with 50'000 dummy rows, so it takes some time for it to be ready.
4. Then the following query can be executed :
- SQLAlchemy container: within the `alchemy-test` container, run `python query.py`, which queries all rows from Event table and prints the number of rows.
- Flask-SQLAlchemy container: make a GET request to `http://localhost:5000/listevents` and it will return the number of rows.
Should be 50'000 in both cases.
5. Regarding the memory consumption now, which can be monitored with `docker stats`:
- SQLAlchemy container: MEM Usage of the query [Before vs During vs After] = [30Mb, 105Mb, 30Mb] so it completely comes back down to the initial value.
- Flask-SQLAlchemy container: MEM Usage of the query [Before vs During vs After] = [78Mb, 156Mb, 86Mb], and it never comes back down to the initial value, unless I hit save in a flask files to restart the container. Here, 8Mb is okay, but in my actual use case on AWS, I don't have much RAM available, the queries I have to make are larger than this, resulting in my RAM staying overwhelmed because of that problem, and significantly slowing down everything.
When I run the same experiment with my Flask-SQLAlchemy container, but removing the polymorphic association between the tables and simply having one table that unites columns from both tables, so the table :
```
class Event(db.Model):
__tablename__ = 'Event'
id = db.Column(db.Integer, primary_key=True)
notebook_id = db.Column(db.String(100), nullable=False)
event_type = db.Column(db.String(32), nullable=False)
col_1 = db.Column(db.String(200), nullable=False)
```
and populating the table with the same content and as many rows, I observe that the RAM comes back to the initial consumption as I would expect after the route responds.
So I'm not completely sure whether this a bug between Flask-SQLAlchemy and the polymorphic association or whether there's something wrong with my code. I tried to keep the implementation as simple as possible and follow the documentation for the polymorphic relationship, which makes me believe there's something strange happening here.
Let me know if you need any clarification or any additional context, thank you for your help !
## Context
<!--Complete the following for context, and add any other relevant context-->
As listed in the `requirements.txt` and in the `Dockerfile` :
- Python version: 3.9.16
- Flask-SQLAlchemy version: 3.0.3
- SQLAlchemy version: 2.0.9
|
closed
|
2023-12-15T14:58:10Z
|
2023-12-30T00:56:35Z
|
https://github.com/pallets-eco/flask-sqlalchemy/issues/1289
|
[] |
Rmarieta
| 1
|
Teemu/pytest-sugar
|
pytest
| 170
|
test failure with pytest 4.2
|
Caused by https://github.com/pytest-dev/pytest/commit/0f546c4670146fbb89407cad85518e3a7dcfa833 (pytest 4.2.0):
The test itself works in pytest without pytest-sugar, so likely caused by overridden hooks.
```
% pytest -k test_collect_error
Test session starts (platform: linux, Python 3.7.2, pytest 4.2.1.dev64+g67dd10de, pytest-sugar 0.9.2)
rootdir: …/Vcs/pytest-sugar, inifile:
plugins: sugar-0.9.2
collecting ...
――――――――――――――――――――――― TestTerminalReporter.test_collect_error ―――――――――――――――――――――――
self = <test_sugar.TestTerminalReporter object at 0x7f8d69310278>
testdir = <Testdir local('/tmp/pytest-of-user/pytest-773/test_collect_error0')>
def test_collect_error(self, testdir):
testdir.makepyfile("""raise ValueError(0)""")
assert_count(testdir)
result = testdir.runpytest('--force-sugar')
result.stdout.fnmatch_lines([
"*ERROR collecting test_collect_error.py*",
"test_collect_error.py:1: in <module>",
" raise ValueError(0)",
> "E ValueError: 0",
])
E Failed: nomatch: '*ERROR collecting test_collect_error.py*'
E and: 'Test session starts (platform: linux, Python 3.7.2, pytest 4.2.1.dev64+g67dd10de, pytest-sugar 0.9.2)'
E and: 'rootdir: /tmp/pytest-of-user/pytest-773/test_collect_error0, inifile:'
E and: 'plugins: sugar-0.9.2'
E and: '\r'
E and: '―――――――――――――――――――――――――――― test_collect_error.py ―――――――――――――――――――――――――――――'
E and: 'test_collect_error.py:1: in <module>'
E and: ' raise ValueError(0)'
E and: 'E ValueError: 0'
E and: ''
E and: '!!!!!!!!!!!!!!!!!!! Interrupted: 1 errors during collection !!!!!!!!!!!!!!!!!!!!'
E and: ''
E and: 'Results (0.04s):'
E and: ''
E remains unmatched: '*ERROR collecting test_collect_error.py*'
…/Vcs/pytest-sugar/test_sugar.py:430: Failed
-------------------------------- Captured stdout call ---------------------------------
============================= test session starts ==============================
platform linux -- Python 3.7.2, pytest-4.2.1.dev64+g67dd10de, py-1.7.0, pluggy-0.8.1
rootdir: /tmp/pytest-of-user/pytest-773/test_collect_error0, inifile:
collected 0 items / 1 errors
==================================== ERRORS ====================================
____________________ ERROR collecting test_collect_error.py ____________________
test_collect_error.py:1: in <module>
raise ValueError(0)
E ValueError: 0
!!!!!!!!!!!!!!!!!!! Interrupted: 1 errors during collection !!!!!!!!!!!!!!!!!!!!
=========================== 1 error in 0.06 seconds ============================
Test session starts (platform: linux, Python 3.7.2, pytest 4.2.1.dev64+g67dd10de, pytest-sugar 0.9.2)
rootdir: /tmp/pytest-of-user/pytest-773/test_collect_error0, inifile:
plugins: sugar-0.9.2
―――――――――――――――――――――――――――― test_collect_error.py ―――――――――――――――――――――――――――――
test_collect_error.py:1: in <module>
raise ValueError(0)
E ValueError: 0
!!!!!!!!!!!!!!!!!!! Interrupted: 1 errors during collection !!!!!!!!!!!!!!!!!!!!
Results (0.05s):
Test session starts (platform: linux, Python 3.7.2, pytest 4.2.1.dev64+g67dd10de, pytest-sugar 0.9.2)
rootdir: /tmp/pytest-of-user/pytest-773/test_collect_error0, inifile:
plugins: sugar-0.9.2
―――――――――――――――――――――――――――― test_collect_error.py ―――――――――――――――――――――――――――――
test_collect_error.py:1: in <module>
raise ValueError(0)
E ValueError: 0
!!!!!!!!!!!!!!!!!!! Interrupted: 1 errors during collection !!!!!!!!!!!!!!!!!!!!
Results (0.04s):
test_sugar.py ⨯ 100% ██████████
Results (0.25s):
1 failed
- test_sugar.py:422 TestTerminalReporter.test_collect_error
27 deselected
```
_Originally posted by @blueyed in https://github.com/Frozenball/pytest-sugar/issues/167#issuecomment-462462930_
|
closed
|
2019-02-11T20:27:53Z
|
2019-06-09T10:03:14Z
|
https://github.com/Teemu/pytest-sugar/issues/170
|
[] |
blueyed
| 2
|
robotframework/robotframework
|
automation
| 4,494
|
Enhance public `robot.api.Languages` and `robot.api.Language` APIs
|
In order to support languages reasonably in RobotCode (and other tools), it would be nice if we could talk about the Language API again.
1. To give the user a good error message which language or custom language was not loaded or found, it should be possible to load individual languages and import custom language files.
- In the current implementation I can either create the `Languages` class in the hope that it works, in case of error I can not easily filter out the language that does not work.
- `add_language` can't load user defined language files
2. There should be a public API that returns all available languages.
- already exists, is just not public
3. The process of loading custom language files should be separated from loading the real language.
- Before loading the real languages, the custom language files should be loaded, only then you can load the language
- I think this should also be done with the command line parameters, there should be 2 arguments, one for loading the custom language files and the current one for the real language
4. it should be possible to create a `Languages` instance that does not insert English and the deprecated single forms of the headers automatically
- I currently have to filter it out again if I want to show only the specified languages e.g. in the completion.
Let's discuss ;-)
|
closed
|
2022-10-03T23:59:47Z
|
2022-10-11T19:26:43Z
|
https://github.com/robotframework/robotframework/issues/4494
|
[
"priority: medium",
"task",
"rc 2",
"acknowledge"
] |
d-biehl
| 23
|
apache/airflow
|
automation
| 47,988
|
Do not call runtime-checks api from the task sdk anymore
|
### Body
https://github.com/apache/airflow/pull/47893 removed the significance of doing: `/task_instances/{uuid}/runtime-checks` during task runs. We did it earlier to register asset changes but that has fundamentally changed in https://github.com/apache/airflow/pull/47677.
Since that API doesnt do much, it would be nice to just remove the API as well as its consumption.
### Committer
- [x] I acknowledge that I am a maintainer/committer of the Apache Airflow project.
|
open
|
2025-03-20T07:57:56Z
|
2025-03-21T09:21:52Z
|
https://github.com/apache/airflow/issues/47988
|
[
"area:API",
"kind:meta",
"priority:medium",
"area:task-execution-interface-aip72",
"area:task-sdk"
] |
amoghrajesh
| 2
|
miguelgrinberg/python-socketio
|
asyncio
| 694
|
Question: How to run the socketio client?
|
Hello there! I built a chat app using Flask-SocketIO and I am trying to build a bot for it. I decided to use this module for the client module. I needed it for real-time sending and receiving. Now I am at the part where I need to run it. I couldn't find any information on the docs.
My Bot Code:
```python
import socketio
class Bot(object):
_CHAT_URL = "<my flask-socketio server>"
room_code = None
def __init__(self, bot_username):
self.bot_username = bot_username
self.sio = socketio.Client()
self.sio.connect(self._CHAT_URL)
self.sio.on('message', self._on_message)
def _on_message(self, **kwargs):
if not self.room_code:
raise Exception("Room Code Variable (room_code) not found.")
self.on_message(**kwargs)
def on_message(self, json):
pass
def send_message(self, message_json):
self.sio.emit('message', message_json)
def run(self):
pass
```
So my question, how do I run the client from the run method?
|
closed
|
2021-05-30T20:25:04Z
|
2021-05-30T22:28:35Z
|
https://github.com/miguelgrinberg/python-socketio/issues/694
|
[
"question"
] |
TechStudent10
| 6
|
graphistry/pygraphistry
|
jupyter
| 301
|
[BUG] `CONTAINS(...)` filters cause server error if NULL values
|
**Describe the bug**
Applying a `CONTAINS(some_attribute, "some_substring")` filter to an attribute that contains NULL values (`None` or `np.nan` in the dataframes used for graph creation) causes a (presumed) internal server error - UI shows `Herding stray GPUs...` then requests to reload.
**To Reproduce**
Example graph: https://hub.graphistry.com/graph/graph.html?dataset=cb1d727659684c74bb64195e1881d1f9
Working example: filter with `CONTAINS(edge:attribute_1, "foo") -> works as expected
Breaking example: filter with `CONTAINS(point:attribute_1, "a") -> `Herding stray GPUs...`
**Expected behavior**
Filtering using `CONTAINS()` with NULL values in attribute should just filter out those NULL values and not break.
**Actual behavior**
`Herding stray GPUs`
**Screenshots**
See graph example above.
**Browser environment (please complete the following information):**
- OS: PopOS
- Browser: Firefox
**Graphistry GPU server environment**
- Graphistry hosted
|
closed
|
2022-01-21T20:16:15Z
|
2022-01-22T09:15:24Z
|
https://github.com/graphistry/pygraphistry/issues/301
|
[
"bug",
"p3"
] |
DBCerigo
| 2
|
jumpserver/jumpserver
|
django
| 14,314
|
[Question] nginx二级目录代理 jms 无法访问
|
### 产品版本
jumpserver/jms_all:latest
### 版本类型
- [X] 社区版
- [ ] 企业版
- [ ] 企业试用版
### 安装方式
- [ ] 在线安装 (一键命令安装)
- [ ] 离线包安装
- [X] All-in-One
- [ ] 1Panel
- [ ] Kubernetes
- [ ] 源码安装
### 环境信息
docker部署
### 🤔 问题描述
nginx二级目录代理 jms 无法访问,具体配置如下:

### 期望结果
nginx二级目录代理 jms 如何配置?
### 补充信息
_No response_
|
closed
|
2024-10-16T14:03:49Z
|
2024-11-28T03:26:33Z
|
https://github.com/jumpserver/jumpserver/issues/14314
|
[
"🤔 Question"
] |
caichangchun
| 3
|
pytorch/vision
|
computer-vision
| 8,255
|
`to_image` does not handle numpy 2D arrays
|
### 🐛 Describe the bug
[`to_image`](https://github.com/pytorch/vision/blob/806dba678d5b01f6e8a46f7c48fdf8c09369a267/torchvision/transforms/v2/functional/_type_conversion.py#L11) should be able to handle [numpy arrays](https://numpy.org/doc/stable/reference/generated/numpy.array.html) with shape `(H, W)`. This corresponds to the previous behavior of [`to_tensor`](https://github.com/pytorch/vision/blob/806dba678d5b01f6e8a46f7c48fdf8c09369a267/torchvision/transforms/functional.py#L149). Running the following:
```python
import numpy as np
from torchvision.transforms.v2.functional import to_image
img_npy = np.random.randint(0, 256, (224, 224), dtype=np.uint8)
to_image(img_npy)
```
results in error:
```
Traceback (most recent call last):
File "<stdin>", line 1, in <module>
File "/home/mantasu/programs/anaconda/envs/glasses-detector-312/lib/python3.12/site-packages/torchvision/transforms/v2/functional/_type_conversion.py", line 14, in to_image
output = torch.from_numpy(inpt).permute((2, 0, 1)).contiguous()
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
RuntimeError: permute(sparse_coo): number of dimensions in the tensor input does not match the length of the desired ordering of dimensions i.e. input.dim() = 2 is not equal to len(dims) = 3
```
PIL grayscale images are handled correctly:
```python
from PIL import Image
img_pil = Image.fromarray(img_npy)
print(to_image(img_pil).shape) # (1, 224, 224)
```
### Versions
```
PyTorch version: 2.2.0
Is debug build: False
CUDA used to build PyTorch: 12.1
ROCM used to build PyTorch: N/A
OS: Ubuntu 22.04.3 LTS (x86_64)
GCC version: (Ubuntu 11.4.0-1ubuntu1~22.04) 11.4.0
Clang version: Could not collect
CMake version: Could not collect
Libc version: glibc-2.35
Python version: 3.11.7 | packaged by conda-forge | (main, Dec 23 2023, 14:43:09) [GCC 12.3.0] (64-bit runtime)
Python platform: Linux-5.15.133.1-microsoft-standard-WSL2-x86_64-with-glibc2.35
Is CUDA available: True
CUDA runtime version: Could not collect
CUDA_MODULE_LOADING set to: LAZY
GPU models and configuration: GPU 0: NVIDIA GeForce RTX 3080 Ti Laptop GPU
Nvidia driver version: 546.33
cuDNN version: Probably one of the following:
/usr/lib/x86_64-linux-gnu/libcudnn.so.8.9.7
/usr/lib/x86_64-linux-gnu/libcudnn_adv_infer.so.8.9.7
/usr/lib/x86_64-linux-gnu/libcudnn_adv_train.so.8.9.7
/usr/lib/x86_64-linux-gnu/libcudnn_cnn_infer.so.8.9.7
/usr/lib/x86_64-linux-gnu/libcudnn_cnn_train.so.8.9.7
/usr/lib/x86_64-linux-gnu/libcudnn_ops_infer.so.8.9.7
/usr/lib/x86_64-linux-gnu/libcudnn_ops_train.so.8.9.7
HIP runtime version: N/A
MIOpen runtime version: N/A
Is XNNPACK available: True
CPU:
Architecture: x86_64
CPU op-mode(s): 32-bit, 64-bit
Address sizes: 39 bits physical, 48 bits virtual
Byte Order: Little Endian
CPU(s): 24
On-line CPU(s) list: 0-23
Vendor ID: GenuineIntel
Model name: 12th Gen Intel(R) Core(TM) i7-12800HX
CPU family: 6
Model: 151
Thread(s) per core: 2
Core(s) per socket: 12
Socket(s): 1
Stepping: 2
BogoMIPS: 4607.99
Flags: fpu vme de pse tsc msr pae mce cx8 apic sep mtrr pge mca cmov pat pse36 clflush mmx fxsr sse sse2 ss ht syscall nx pdpe1gb rdtscp lm constant_tsc rep_good nopl xtopology tsc_reliable nonstop_tsc cpuid pni pclmulqdq vmx ssse3 fma cx16 sse4_1 sse4_2 x2apic movbe popcnt tsc_deadline_timer aes xsave avx f16c rdrand hypervisor lahf_lm abm 3dnowprefetch ssbd ibrs ibpb stibp ibrs_enhanced tpr_shadow vnmi ept vpid ept_ad fsgsbase tsc_adjust bmi1 avx2 smep bmi2 erms invpcid rdseed adx smap clflushopt clwb sha_ni xsaveopt xsavec xgetbv1 xsaves avx_vnni umip waitpkg gfni vaes vpclmulqdq rdpid movdiri movdir64b fsrm serialize flush_l1d arch_capabilities
Virtualization: VT-x
Hypervisor vendor: Microsoft
Virtualization type: full
L1d cache: 576 KiB (12 instances)
L1i cache: 384 KiB (12 instances)
L2 cache: 15 MiB (12 instances)
L3 cache: 25 MiB (1 instance)
Vulnerability Gather data sampling: Not affected
Vulnerability Itlb multihit: Not affected
Vulnerability L1tf: Not affected
Vulnerability Mds: Not affected
Vulnerability Meltdown: Not affected
Vulnerability Mmio stale data: Not affected
Vulnerability Retbleed: Mitigation; Enhanced IBRS
Vulnerability Spec rstack overflow: Not affected
Vulnerability Spec store bypass: Mitigation; Speculative Store Bypass disabled via prctl and seccomp
Vulnerability Spectre v1: Mitigation; usercopy/swapgs barriers and __user pointer sanitization
Vulnerability Spectre v2: Mitigation; Enhanced IBRS, IBPB conditional, RSB filling, PBRSB-eIBRS SW sequence
Vulnerability Srbds: Not affected
Vulnerability Tsx async abort: Not affected
Versions of relevant libraries:
[pip3] numpy==1.26.3
[pip3] torch==2.2.0
[pip3] torchaudio==2.2.0
[pip3] torchvision==0.17.0
[pip3] triton==2.2.0
[conda] blas 1.0 mkl conda-forge
[conda] ffmpeg 4.3 hf484d3e_0 pytorch
[conda] libjpeg-turbo 2.0.0 h9bf148f_0 pytorch
[conda] mkl 2023.1.0 h213fc3f_46344
[conda] numpy 1.26.3 py311h64a7726_0 conda-forge
[conda] pytorch 2.2.0 py3.11_cuda12.1_cudnn8.9.2_0 pytorch
[conda] pytorch-cuda 12.1 ha16c6d3_5 pytorch
[conda] pytorch-mutex 1.0 cuda pytorch
[conda] torchaudio 2.2.0 py311_cu121 pytorch
[conda] torchtriton 2.2.0 py311 pytorch
[conda] torchvision 0.17.0 py311_cu121 pytorch
```
|
closed
|
2024-02-05T14:27:58Z
|
2024-02-06T11:10:20Z
|
https://github.com/pytorch/vision/issues/8255
|
[] |
mantasu
| 0
|
mljar/mercury
|
jupyter
| 143
|
Import class from another directory that notebook
|
Hello,
I have python project with notebooks.
Noteboks are in direcotry ./notebooks
Python classes and code are in ./twitter folder.
When i want to use classes in notebooks and use in Mercury app i have error
ModuleNotFoundError: No module named 'twitter'
I use: from twitter ....
So it look like mercury cannot access my local folder/module.
I found solution to add my code to conda modules but i need create that module but maybe is another way
to show mercury that i have local code ?
Thanks for answer.
|
closed
|
2022-07-14T10:33:17Z
|
2022-07-14T12:08:59Z
|
https://github.com/mljar/mercury/issues/143
|
[] |
kamiljaszczynski
| 3
|
autokey/autokey
|
automation
| 135
|
Autokey installs but won't run on (18.04)
|
## Classification:
Bug
## Reproducibility:
- Try to install autokey through ubuntu store
- Try to install autokey with the pip3 setup.py install command
- Try to install autokey with the apt-get autokey-gtk
## Summary
I am trying to install the autokey package on Ubuntu 18.04 and It worked the first time but when I had to remove it I couldn't get it to reinstall again.
## Steps to Reproduce
Remove the program the following way:
- sudo apt-get purge --auto-remove autokey-gtk
Install the program by using one of the following ways;
- sudo apt-get update;
sudo apt-get autokey-gtk
- pip3 setup.py install;
## Expected Results
When i try to open autokey from terminal or the pacakge explorere I would expect a GUI to open.
## Actual Results
The program won't open the GUI.
## Error While loading
autokey-gtk --version
Traceback (most recent call last):
File "/usr/local/bin/autokey-gtk", line 11, in <module>
load_entry_point('autokey==0.94.0', 'console_scripts', 'autokey-gtk')()
File "/home/ricks/.local/lib/python3.6/site-packages/pkg_resources/__init__.py", line 480, in load_entry_point
return get_distribution(dist).load_entry_point(group, name)
File "/home/ricks/.local/lib/python3.6/site-packages/pkg_resources/__init__.py", line 2693, in load_entry_point
return ep.load()
File "/home/ricks/.local/lib/python3.6/site-packages/pkg_resources/__init__.py", line 2324, in load
return self.resolve()
File "/home/ricks/.local/lib/python3.6/site-packages/pkg_resources/__init__.py", line 2330, in resolve
module = __import__(self.module_name, fromlist=['__name__'], level=0)
File "/usr/local/lib/python3.6/dist-packages/autokey-0.94.0-py3.6.egg/autokey/gtkui/__main__.py", line 4, in <module>
from autokey.gtkapp import Application
File "/usr/local/lib/python3.6/dist-packages/autokey-0.94.0-py3.6.egg/autokey/gtkapp.py", line 44, in <module>
from autokey import service, monitor
File "/usr/local/lib/python3.6/dist-packages/autokey-0.94.0-py3.6.egg/autokey/service.py", line 25, in <module>
from .iomediator.key import Key
ModuleNotFoundError: No module named 'autokey.iomediator'
## my thoughts
I am new to Linux so I accidentally removed some crucial files when purging the program.
## Version
AutoKey-Py3 version: Latest version
Installed via: (PPA, pip and ubuntu store)
Distro: Ubuntu Bionic beaver (18.04)
|
closed
|
2018-04-26T08:00:27Z
|
2018-04-29T04:19:59Z
|
https://github.com/autokey/autokey/issues/135
|
[] |
rickstaa
| 4
|
elliotgao2/toapi
|
api
| 51
|
Ip proxies.
|
closed
|
2017-12-11T01:53:04Z
|
2017-12-13T03:44:28Z
|
https://github.com/elliotgao2/toapi/issues/51
|
[] |
elliotgao2
| 0
|
|
coqui-ai/TTS
|
pytorch
| 3,617
|
[Bug] atttributeError: 'Tacotron2' object has no attribute 'args'
|
### Describe the bug
Traceback (most recent call last):
File "/home/mariah/.local/bin/tts", line 8, in <module>
sys.exit(main())
^^^^^^
File "/home/mariah/Applications/tts/TTS/bin/synthesize.py", line 468, in main
wav = synthesizer.tts(
^^^^^^^^^^^^^^^^
File "/home/mariah/Applications/tts/TTS/utils/synthesizer.py", line 463, in tts
outputs = transfer_voice(
^^^^^^^^^^^^^^^
File "/home/mariah/Applications/tts/TTS/tts/utils/synthesis.py", line 319, in transfer_voice
reference_wav, sr=model.args.encoder_sample_rate if model.args.encoder_sample_rate else model.ap.sample_rate
^^^^^^^^^^
File "/home/mariah/.local/pipx/venvs/tts/lib/python3.11/site-packages/torch/nn/modules/module.py", line 1688, in __getattr__
raise AttributeError(f"'{type(self).__name__}' object has no attribute '{name}'")
AttributeError: 'Tacotron2' object has no attribute 'args'
### To Reproduce
<pre>\ 9-year-old\ Doctor\ Who\ fan\ \[VS0iFWwvDbs\].wav --text "test" --pipe_out --out_path t.wav > error.txt
### Expected behavior
_No response_
### Logs
_No response_
### Environment
```shell
I tried running collect_env_info.py but it says no module found torch
```
### Additional context
_No response_
|
closed
|
2024-03-01T19:40:43Z
|
2025-01-03T09:47:59Z
|
https://github.com/coqui-ai/TTS/issues/3617
|
[
"bug",
"wontfix"
] |
MariahWest01
| 1
|
huggingface/datasets
|
pytorch
| 7,215
|
Iterable dataset map with explicit features causes slowdown for Sequence features
|
### Describe the bug
When performing map, it's nice to be able to pass the new feature type, and indeed required by interleave and concatenate datasets.
However, this can cause a major slowdown for certain types of array features due to the features being re-encoded.
This is separate to the slowdown reported in #7206
### Steps to reproduce the bug
```
from datasets import Dataset, Features, Array3D, Sequence, Value
import numpy as np
import time
features=Features(**{"array0": Sequence(feature=Value("float32"), length=-1), "array1": Sequence(feature=Value("float32"), length=-1)})
dataset = Dataset.from_dict({f"array{i}": [np.zeros((x,), dtype=np.float32) for x in [5000,10000]*25] for i in range(2)}, features=features)
```
```
ds = dataset.to_iterable_dataset()
ds = ds.with_format("numpy").map(lambda x: x)
t0 = time.time()
for ex in ds:
pass
t1 = time.time()
```
~1.5 s on main
```
ds = dataset.to_iterable_dataset()
ds = ds.with_format("numpy").map(lambda x: x, features=features)
t0 = time.time()
for ex in ds:
pass
t1 = time.time()
```
~ 3 s on main
### Expected behavior
I'm not 100% sure whether passing new feature types to formatted outputs of map should be supported or not, but assuming it should, then there should be a cost-free way to specify the new feature type - knowing feature type is required by interleave_datasets and concatenate_datasets for example
### Environment info
3.0.2
|
open
|
2024-10-10T22:08:20Z
|
2024-10-10T22:10:32Z
|
https://github.com/huggingface/datasets/issues/7215
|
[] |
alex-hh
| 0
|
Textualize/rich
|
python
| 2,921
|
Add comparison methods to rich Text
|
Text could use comparison methods for this use case: https://github.com/Textualize/textual/issues/2261
Comparisons should treat strings and `Text` in the same way. This will allow sorting of `Text` objects.
|
open
|
2023-04-12T10:08:50Z
|
2023-04-12T11:58:26Z
|
https://github.com/Textualize/rich/issues/2921
|
[
"Needs triage"
] |
willmcgugan
| 3
|
lepture/authlib
|
flask
| 204
|
AttributeError: 'NoneType' object has no attribute 'split' if self.scope is None
|
`AttributeError` occurs when the client scopes is `None`
**Error Stacks**
```
def get_allowed_scope(self, scope):
if not scope:
return ''
> allowed = set(self.scope.split())
E AttributeError: 'NoneType' object has no attribute 'split'
```
As informed in [rfc6749#section-3.3](https://tools.ietf.org/html/rfc6749#section-3.3), that
```
...
The authorization server MAY fully or partially ignore the scope
requested by the client, based on the authorization server policy or
the resource owner's instructions.
...
```
I think its better if we add empty string as default value:
```python
...
# line 68
@property
def scope(self):
return self.client_metadata.get('scope', '')
...
```
All tests run well
Pull request: [github.com/lepture/authlib/pull/205](https://github.com/lepture/authlib/pull/205)
|
closed
|
2020-03-12T14:52:46Z
|
2020-03-20T03:22:10Z
|
https://github.com/lepture/authlib/issues/204
|
[
"bug"
] |
GulaAren
| 0
|
huggingface/datasets
|
machine-learning
| 7,443
|
index error when num_shards > len(dataset)
|
In `ds.push_to_hub()` and `ds.save_to_disk()`, `num_shards` must be smaller than or equal to the number of rows in the dataset, but currently this is not checked anywhere inside these functions. Attempting to invoke these functions with `num_shards > len(dataset)` should raise an informative `ValueError`.
I frequently work with datasets with a small number of rows where each row is pretty large, so I often encounter this issue, where the function runs until the shard index in `ds.shard(num_shards, indx)` goes out of bounds. Ideally, a `ValueError` should be raised before reaching this point (i.e. as soon as `ds.push_to_hub()` or `ds.save_to_disk()` is invoked with `num_shards > len(dataset)`).
It seems that adding something like:
```python
if len(self) < num_shards:
raise ValueError(f"num_shards ({num_shards}) must be smaller than or equal to the number of rows in the dataset ({len(self)}). Please either reduce num_shards or increase max_shard_size to make sure num_shards <= len(dataset).")
```
to the beginning of the definition of the `ds.shard()` function [here](https://github.com/huggingface/datasets/blob/f693f4e93aabafa878470c80fd42ddb10ec550d6/src/datasets/arrow_dataset.py#L4728) would deal with this issue for both `ds.push_to_hub()` and `ds.save_to_disk()`, but I'm not exactly sure if this is the best place to raise the `ValueError` (it seems that a more correct way to do it would be to write separate checks for `ds.push_to_hub()` and `ds.save_to_disk()`). I'd be happy to submit a PR if you think something along these lines would be acceptable.
|
open
|
2025-03-10T22:40:59Z
|
2025-03-10T23:43:08Z
|
https://github.com/huggingface/datasets/issues/7443
|
[] |
eminorhan
| 1
|
gradio-app/gradio
|
machine-learning
| 10,708
|
Cache especific examples
|
Can I cache only some examples from the list? Scenario: I have some examples that can only be run locally on a good GPU and others that can be run on huggingface's Zero GPU.
I thought of something like "example_labels" something like "examples_enable_cache" = [True, False, True, False ....]
|
open
|
2025-03-02T14:25:38Z
|
2025-03-03T14:10:47Z
|
https://github.com/gradio-app/gradio/issues/10708
|
[
"enhancement"
] |
elismasilva
| 1
|
django-cms/django-cms
|
django
| 7,449
|
KeyError at /xxxxx - After removing a plugin
|
<!--
Please fill in each section below, otherwise, your issue will be closed.
This info allows django CMS maintainers to diagnose (and fix!) your issue
as quickly as possible.
-->
## Description
I have a plugin **NavbarPlugin** that I coded inside **cms_plugins.py.**
- The code :
```py
@plugin_pool.register_plugin
class NavbarPlugin(CMSPluginBase):
model = CMSPlugin
render_template = "frontend/inviso_plugins_templates/menu/inviso_navbar.html"
name="Navbar plugin"
module = "Inviso"
def render(self, context, instance, placeholder):
context = super().render(context, instance, placeholder)
return context
```
I used this plugin inside a page that I created, then it worked well.
Then I decided to delete the plugin from the code inside **cms_plugins.py**, but I forgot to remove it from the page in the first place.
Then I got an error like :
```
KeyError at /actualites/
'NavbarPlugin'
```
I decided then to undo the suppression of the plugin from **cms_plugins.py**, but then it turned out that there's always the same error again and again. I searched on the internet and all but I didn't got what I searched for.
Thanks
<!--
If this is a security issue stop immediately and follow the instructions at:
http://docs.django-cms.org/en/latest/contributing/development-policies.html#reporting-security-issues
-->
## Steps to reproduce
<!--
Clear steps describing how to reproduce the issue.
Steps to reproduce the behavior:
1. Go to '...'
2. Click on '....'
3. Scroll down to '....'
4. See error
-->
- Create a plugin in **cms_plugins.py**
- Use the plugin in a page
- Delete the plugin code in **cms_plugins.py**
- Refresh the page (or the project)
- Getting an error
- Rewritting the same plugin code in **cms_plugins.py**
- Refresh the page (or the project)
## Expected behaviour
<!--
A clear and concise description of what you expected to happen.
-->
After rewritting the plugin code in **cms_plugins.py**, the plugin should be seen by the application and no errors should occur anymore.
## Actual behaviour
The same error that occurs after deleting the plugin code from **cms_plugins.py** persists
<!--
A clear and concise description of what is actually happening.
-->
## Screenshots
<!--If applicable, add screenshots to help explain your problem.
-->
## Additional information (CMS/Python/Django versions)
<!--
Add any other context about the problem such as environment,
CMS/Python/Django versions, logs etc. here.
-->
## Do you want to help fix this issue?
<!--
The django CMS project is managed and kept alive by its open source community and is backed by the [django CMS Association](https://www.django-cms.org/en/about-us/). We therefore welcome any help and are grateful if people contribute to the project. Please use 'x' to check the items below.
-->
* [ ] Yes, I want to help fix this issue and I will join #workgroup-pr-review on [Slack](https://www.django-cms.org/slack) to confirm with the community that a PR is welcome.
* [ ] No, I only want to report the issue.
|
closed
|
2022-12-07T13:51:20Z
|
2023-01-11T07:07:17Z
|
https://github.com/django-cms/django-cms/issues/7449
|
[
"status: marked for rejection"
] |
cedricrabarijohn
| 2
|
zappa/Zappa
|
django
| 655
|
[Migrated] module 'pip' has no attribute 'get_installed_distributions'
|
Originally from: https://github.com/Miserlou/Zappa/issues/1658 by [nabaz](https://github.com/nabaz)
<!--- Provide a general summary of the issue in the Title above -->
## module 'pip' has no attribute 'get_installed_distributions' no matter what you do you get this error, so frustrating, makes you not use zappa anymore it's unfortunate
<!--- Provide a more detailed introduction to the issue itself, and why you consider it to be a bug -->
<!--- Also, please make sure that you are running Zappa _from a virtual environment_ and are using Python 2.7/3.6 -->
3.6
## Expected Behavior
<!--- Tell us what should happen -->
module 'pip' has no attribute 'get_installed_distributions'
## Actual Behavior
<!--- Tell us what happens instead -->
## Possible Fix
<!--- Not obligatory, but suggest a fix or reason for the bug -->
## Steps to Reproduce
<!--- Provide a link to a live example, or an unambiguous set of steps to -->
<!--- reproduce this bug include code to reproduce, if relevant -->
1.
2.
3.
## Your Environment
<!--- Include as many relevant details about the environment you experienced the bug in -->
* Zappa version used:
* Operating System and Python version:
* The output of `pip freeze`:
* Link to your project (optional):
* Your `zappa_settings.py`:
|
closed
|
2021-02-20T12:32:30Z
|
2022-07-16T06:46:47Z
|
https://github.com/zappa/Zappa/issues/655
|
[
"needs-info"
] |
jneves
| 1
|
simple-login/app
|
flask
| 1,002
|
MX setting for custom domain on self hosted instance
|
first - brilliant product, just set up using the self hosting instructions which were straightforward
one thing to note - in the custom domain setup instructions and DNS verification - MX record is meant to point to _mydomain.com_ but this didn't work for me until this was changed _app.mydomain.com_
|
closed
|
2022-05-19T05:13:28Z
|
2022-05-19T05:48:04Z
|
https://github.com/simple-login/app/issues/1002
|
[] |
mascooler
| 1
|
recommenders-team/recommenders
|
deep-learning
| 1,210
|
[ASK]
|
### NPA metrics
Hello! I am testing NAML, NRMS and NPA algorithms for MIND-dataset. I noticed that the metric HIT@1,5,10 has a non-zero value for the first two algorithms, but for NPA it is zero. Can someone explain why this is so? thnx
|
closed
|
2020-10-01T11:57:21Z
|
2020-10-07T08:18:42Z
|
https://github.com/recommenders-team/recommenders/issues/1210
|
[
"help wanted"
] |
Holyland-EG
| 3
|
fastapi/sqlmodel
|
fastapi
| 314
|
`sqlalchemy.Column` parameters are not passed forward when set on `sqlmodel.Field` and a column is provided via `sa_column`
|
### First Check
- [X] I added a very descriptive title to this issue.
- [X] I used the GitHub search to find a similar issue and didn't find it.
- [X] I searched the SQLModel documentation, with the integrated search.
- [X] I already searched in Google "How to X in SQLModel" and didn't find any information.
- [X] I already read and followed all the tutorial in the docs and didn't find an answer.
- [X] I already checked if it is not related to SQLModel but to [Pydantic](https://github.com/samuelcolvin/pydantic).
- [X] I already checked if it is not related to SQLModel but to [SQLAlchemy](https://github.com/sqlalchemy/sqlalchemy).
### Commit to Help
- [X] I commit to help with one of those options 👆
### Example Code
```python
name: str = Field(
sa_column=Column(
String,
# ... other attrs not exposed in sqlmodel.Field today
),
index=True # this is ignored, must be set on `Column` above
)
```
### Description
`sqlmodel.Field` exposes some but not all fields from `sqlalchemy.Column`. This means in some cases it is necessary to provide a `Column` via the `sa_column=` param on `sqlmodel.Field`. However, when this is the case the parameters set on `sqlmodel.Field` are not forwarded to the new `sa_column` object.
I think the expected behavior here is that parameters set on `Field` would be combined with those from the `sa_column` object. Either this or setting a parameter that will be ignored should trigger a warning/exception along the lines of `"You have set index but also provided a sqlalchemy.Column object, index will be ignored"`.
### Operating System
Linux
### Operating System Details
_No response_
### SQLModel Version
0.0.6
### Python Version
3.9.5
### Additional Context
_No response_
|
closed
|
2022-04-26T18:59:26Z
|
2022-11-22T00:12:00Z
|
https://github.com/fastapi/sqlmodel/issues/314
|
[
"question",
"answered"
] |
JLHasson
| 6
|
mwaskom/seaborn
|
data-visualization
| 3,617
|
seaborn scatterplot hue= datetime bug
|
### my issue is: why 2023 can appear in the below figure's legend?
**Please see below codes.**
```
import pandas as pd
import seaborn as sns
from matplotlib import pyplot as plt
df = pd.DataFrame(
{"Time_str": ["2024,10,1", "2024,11,1"],
"x": [1, 2],
"y": [10, 20]
})
df["Datetime"] = pd.to_datetime(df["Time_str"])
df["year"] = df["Datetime"].dt.year
fig, ax = plt.subplots(figsize=(14, 8))
sns.scatterplot(data=df, x="x", y="y", hue="year")
```

>
|
closed
|
2024-01-16T07:16:49Z
|
2024-01-17T02:21:57Z
|
https://github.com/mwaskom/seaborn/issues/3617
|
[] |
juanjuanlovelearning
| 3
|
amidaware/tacticalrmm
|
django
| 1,255
|
Users can see and assign automatisch Policies from Users/Sites/Clients they do not belong to.
|
I would expect that Automation-Policies are limited the same way as Policy-Overview is?
You should not be able to see Policies created from Users who belong only to specific Clients or Sites.
Also it should not be possible to assign them.

|
open
|
2022-08-23T11:17:38Z
|
2022-08-23T18:39:50Z
|
https://github.com/amidaware/tacticalrmm/issues/1255
|
[
"enhancement"
] |
JSuenram
| 1
|
pyeventsourcing/eventsourcing
|
sqlalchemy
| 101
|
How to install with Cassandra?
|
I'm using pip version 9.01 and I tried using python 3.5, pypy 5.8, and python 2.7
But everytime when I try to install eventsourcing with either cassandra or sqlalchemy, it gives me error like the following:
```
$ pip install eventsourcing[cassandra]
zsh: no matches found: eventsourcing[cassandra]
```
Is there a way that I could install eventsourcing with the specified event store?
|
closed
|
2017-06-27T12:51:57Z
|
2017-06-27T20:01:53Z
|
https://github.com/pyeventsourcing/eventsourcing/issues/101
|
[] |
subokita
| 2
|
psf/black
|
python
| 4,573
|
Projeto
|
closed
|
2025-02-02T01:19:49Z
|
2025-02-02T02:46:46Z
|
https://github.com/psf/black/issues/4573
|
[] |
thiagomsa
| 0
|
|
piskvorky/gensim
|
data-science
| 2,804
|
ModuleNotFoundError: No module named 'testfixtures'
|
<!--
**IMPORTANT**:
- Use the [Gensim mailing list](https://groups.google.com/forum/#!forum/gensim) to ask general or usage questions. Github issues are only for bug reports.
- Check [Recipes&FAQ](https://github.com/RaRe-Technologies/gensim/wiki/Recipes-&-FAQ) first for common answers.
Github bug reports that do not include relevant information and context will be closed without an answer. Thanks!
-->
#### Problem description
* running the following code in `/docs/notebooks/howtos/run_doc2vec_imdb.ipynb`:
`from gensim.test.test_doc2vec import ConcatenatedDoc2Vec`

* A quick review of `/gensim/test/test_doc2vec.py` shows the offending import on line#21. I've tried replacing the line with `from fixtures import log_capture`, but with no improvement.
#### Steps/code/corpus to reproduce
run `run_doc2vec_imdb.ipynb`
#### Versions
Linux-4.15.0-91-generic-x86_64-with-Ubuntu-18.04-bionic
Python 3.6.9 (default, Jul 3 2019, 15:36:16)
[GCC 5.4.0 20160609]
NumPy 1.18.3
SciPy 1.4.1
gensim 3.8.1
FAST_VERSION 1
|
open
|
2020-04-23T03:53:21Z
|
2020-08-13T12:24:45Z
|
https://github.com/piskvorky/gensim/issues/2804
|
[
"documentation",
"impact HIGH",
"reach LOW"
] |
bjpcjp
| 8
|
PeterL1n/BackgroundMattingV2
|
computer-vision
| 167
|
Details of the training process
|
你好,关于训练细节,我请教几个问题:
1、训练中数据集的使用顺序依次为:VideoMatte240K、PhotoMatte13K、Distinctions-646,请问为什么要将PhotoMatte13K、Distinctions-646二者分离而不是合并在一起训练?分离的做法是出于什么样的考虑?
2、对于refine 模型的训练过程中,各个阶段(各个数据集上的训练)都是训练到收敛再进入下一个阶段?如果不是的话,那应该训练到什么样的程度才进入下一阶段?
3、在本文中似乎提到AIM和Distinctions-646数据集差异较大,但为什么没有像RVM(你的另一篇论文)那样,将两者结合训练?
谢谢。
|
open
|
2021-12-21T08:01:38Z
|
2021-12-21T17:21:25Z
|
https://github.com/PeterL1n/BackgroundMattingV2/issues/167
|
[] |
li-wenquan
| 1
|
awesto/django-shop
|
django
| 810
|
Support for django 3.0
|
Please support for django 3.0 .
django-cms has already upgraded
|
open
|
2020-05-04T05:04:06Z
|
2020-06-28T15:39:31Z
|
https://github.com/awesto/django-shop/issues/810
|
[] |
pupattan
| 23
|
pinry/pinry
|
django
| 112
|
No way to change password?
|
I'm setting up a private board and want to create a few accounts for friends and then close registrations. I was going to generate passwords which they could then change to something they prefer but as far as I can tell there's no way to change passwords. Am I just missing where that's done?
|
closed
|
2017-06-21T01:43:16Z
|
2019-12-08T19:35:26Z
|
https://github.com/pinry/pinry/issues/112
|
[
"enhancement"
] |
Flamekebab
| 4
|
Lightning-AI/pytorch-lightning
|
deep-learning
| 19,973
|
FSDPPrecision should support 16-true with a loss scaler
|
### Description & Motivation
https://github.com/Lightning-AI/pytorch-lightning/blob/f6fd046552a1504023cb3386a8a0df418a810e4f/src/lightning/fabric/plugins/precision/fsdp.py#L61
What if I want to use fp16 true, but with a loss scaler? This is closer to DeepSpeed's default settings. With FSDP, 16-true, no loss scaler my model doesn't converge. However, with FSDP, 16-true, and a loss scaler (commented out the assert and fixed the typo'ed return scaler instead of return none line) my model converges.
### Pitch
_No response_
### Alternatives
_No response_
### Additional context
_No response_
cc @borda
|
open
|
2024-06-13T02:13:59Z
|
2024-06-13T21:11:12Z
|
https://github.com/Lightning-AI/pytorch-lightning/issues/19973
|
[
"feature",
"needs triage"
] |
zaptrem
| 1
|
plotly/dash
|
jupyter
| 2,788
|
[Feature Request] Supports more flexible and free component type properties
|
Currently, as mentioned in the documentation ( https://dash.plotly.com/component-properties ), only a few valid formats are supported for component elements as attributes, the hope is that dash will support freer component-type property definitions under the hood, including arbitrary depth nesting, to cope with some tree-like data structures.
|
closed
|
2024-03-11T07:38:53Z
|
2024-07-26T13:06:40Z
|
https://github.com/plotly/dash/issues/2788
|
[
"feature"
] |
CNFeffery
| 2
|
strawberry-graphql/strawberry
|
graphql
| 3,158
|
Default values for scalar arguments passed as string
|
When declaring an optional argument with a scalar type, its default value is passed as a string in the resulting schema. This makes Strawberry-declared schemas incompatible with externally connected GraphQL consumers with strict schema checkers, such as Hasura.
The following code:
```python
from typing import Optional
import strawberry
@strawberry.type
class Query:
@strawberry.field
def example(self,
baz: int,
foo: int | None = None ,
bar: int = 10
) -> None:
return None
schema = strawberry.Schema(query=Query)
```
produces the following default values in the schema:
```
...
"defaultValue": null
...
"defaultValue": "null"
...
"defaultValue": "10"
```
<details>
<summary> Schema inspection </summary>
```graphql
{
"data": {
"__schema": {
"queryType": {
"name": "Query"
},
"mutationType": null,
"types": [
{
"kind": "OBJECT",
"name": "Query",
"description": null,
"fields": [
{
"name": "example",
"description": null,
"args": [
{
"name": "baz",
"description": null,
"type": {
"kind": "NON_NULL",
"name": null,
"ofType": {
"kind": "SCALAR",
"name": "Int",
"ofType": null
}
},
"defaultValue": null
},
{
"name": "foo",
"description": null,
"type": {
"kind": "SCALAR",
"name": "Int",
"ofType": null
},
"defaultValue": "null"
},
{
"name": "bar",
"description": null,
"type": {
"kind": "NON_NULL",
"name": null,
"ofType": {
"kind": "SCALAR",
"name": "Int",
"ofType": null
}
},
"defaultValue": "10"
}
```
</details>
## System Information
Python 3.11
Ubuntu 22.10
Strawberry version: 0.209.2
|
closed
|
2023-10-18T09:52:49Z
|
2025-03-20T15:56:26Z
|
https://github.com/strawberry-graphql/strawberry/issues/3158
|
[
"bug"
] |
ichorid
| 7
|
deezer/spleeter
|
tensorflow
| 130
|
[Bug] Tuple formatting incorrectly included in output directory name
|
## Description
I am using the [separator.py](https://github.com/deezer/spleeter/blob/master/spleeter/separator.py) file to include `spleeter` in my own Python development. The [separate_to_file](https://github.com/deezer/spleeter/blob/85ff00797f6c615c62885793923eca952e9e791f/spleeter/separator.py#L93) function is erroneously including parentheses in the name of the output directory. The user does not have a way to avoid this formatting.
Example:
> Input filename: `GilScottHeron_WeAlmostLostDetroit.mp3`
> Output directory name : `('GilScottHeron_WeAlmostLostDetroit', '.mp3')/`
## Step to reproduce
1. Installed using `pip`
2. Run as a Python script
3. Got no error. The output directory name formatting is tuple.
```python
from spleeter.spleeter.separator import Separator
separator = Separator('spleeter:2stems')
filein = 'GilScottHeron_WeAlmostLostDetroit.mp3'
fileout = './stems'
separator.separate_to_file(filein, fileout, codec='mp3')
```
## Output
Output directory name : `./stems/('GilScottHeron_WeAlmostLostDetroit', '.mp3')/`
Expected output: `./stems/GilScottHeron_WeAlmostLostDetroit/`
## Environment
<!-- Fill the following table -->
| | |
| ----------------- | ------------------------------- |
| OS | MacOS |
| Installation type | `pip` |
| RAM available | 8 GB |
| Hardware spec | CPU: 3.2 GHz Intel Core i5, GPU: NVIDIA GeForce GT 755M 1 GB |
## Additional context
The reason for this bug is [line 124 in `separator.py`](https://github.com/deezer/spleeter/blob/85ff00797f6c615c62885793923eca952e9e791f/spleeter/separator.py#L124). There needs to be a `[0]` added after the output of `splitext` so that the directory name is created from a `string`, not a `tuple`.
|
closed
|
2019-11-23T23:27:28Z
|
2019-11-25T14:34:13Z
|
https://github.com/deezer/spleeter/issues/130
|
[
"bug",
"next release"
] |
johnwmillr
| 2
|
MycroftAI/mycroft-core
|
nlp
| 2,425
|
Language Settings not working
|
## Be clear about the software, hardware and version you are running
* I'm running a rapsberrypi 3
* With Mycroft from master branch
* With the standard Wake Word
## Try to provide steps that we can use to replicate the Issue
install mycroft as in the tutortial described
use the command: mycroft-config set lang "de-de"
restart mycroft instance
mycroft is still only working in english and not in german or any other language
|
closed
|
2019-12-13T19:11:30Z
|
2019-12-14T21:00:18Z
|
https://github.com/MycroftAI/mycroft-core/issues/2425
|
[] |
flozi00
| 8
|
adbar/trafilatura
|
web-scraping
| 81
|
Trafilatura duplicating text
|
Issue described [here](https://stackoverflow.com/questions/67812700/has-anyone-experience-issues-with-duplicated-text-while-using-trafilatura)
|
closed
|
2021-06-03T13:31:37Z
|
2021-06-04T17:12:28Z
|
https://github.com/adbar/trafilatura/issues/81
|
[
"bug"
] |
cta2106
| 8
|
iperov/DeepFaceLab
|
deep-learning
| 820
|
Is OpenCL supported ?
|
I have an AMD videocard and thus i cannot run the proprietary cuda. Is it possible or be possible in the future to use OpenCL ?
|
open
|
2020-07-08T20:01:17Z
|
2023-06-08T22:24:52Z
|
https://github.com/iperov/DeepFaceLab/issues/820
|
[] |
ghost
| 3
|
plotly/dash-table
|
dash
| 622
|
Table export to xlsx, applying styles
|
It would be amazing to be able to export the table including the formats applied to the dash table. This was already hinted at here:
https://github.com/plotly/dash-table/issues/313#issuecomment-503628712
|
open
|
2019-10-09T05:16:48Z
|
2022-09-20T08:14:46Z
|
https://github.com/plotly/dash-table/issues/622
|
[
"dash-type-enhancement"
] |
bjonen
| 3
|
bmoscon/cryptofeed
|
asyncio
| 738
|
Do XX on websocket error?
|
I am listening to bybit websocket and doing operations on trades
>f = FeedHandler()
f.add_feed(BYBIT, channels=[TRADES], symbols=['BTC-USD-PERP'], callbacks={TRADES: trade}, timeout=-1)
f.run()
Whenever the live wss goes down, I get this expected trackback error.
>WARNING : BYBIT.ws.1: encountered connection issue no close frame received or sent - reconnecting in 1.0 seconds...
Traceback (most recent call last):
File "/home/ubuntu/.local/lib/python3.8/site-packages/websockets/legacy/protocol.py", line 944, in transfer_data
message = await self.read_message()
File "/home/ubuntu/.local/lib/python3.8/site-packages/websockets/legacy/protocol.py", line 1013, in read_message
frame = await self.read_data_frame(max_size=self.max_size)
File "/home/ubuntu/.local/lib/python3.8/site-packages/websockets/legacy/protocol.py", line 1089, in read_data_frame
frame = await self.read_frame(max_size)
File "/home/ubuntu/.local/lib/python3.8/site-packages/websockets/legacy/protocol.py", line 1144, in read_frame
frame = await Frame.read(
File "/home/ubuntu/.local/lib/python3.8/site-packages/websockets/legacy/framing.py", line 70, in read
data = await reader(2)
File "/usr/lib/python3.8/asyncio/streams.py", line 723, in readexactly
await self._wait_for_data('readexactly')
File "/usr/lib/python3.8/asyncio/streams.py", line 517, in _wait_for_data
await self._waiter
ConnectionResetError: [Errno 104] Connection reset by peer
>
>The above exception was the direct cause of the following exception:
>
>Traceback (most recent call last):
File "/home/ubuntu/.local/lib/python3.8/site-packages/cryptofeed/connection_handler.py", line 69, in _create_connection
await self._handler(connection, self.handler)
File "/home/ubuntu/.local/lib/python3.8/site-packages/cryptofeed/connection_handler.py", line 115, in _handler
async for message in connection.read():
File "/home/ubuntu/.local/lib/python3.8/site-packages/cryptofeed/connection.py", line 320, in read
async for data in self.conn:
File "/home/ubuntu/.local/lib/python3.8/site-packages/websockets/legacy/protocol.py", line 481, in __aiter__
yield await self.recv()
File "/home/ubuntu/.local/lib/python3.8/site-packages/websockets/legacy/protocol.py", line 552, in recv
await self.ensure_open()
File "/home/ubuntu/.local/lib/python3.8/site-packages/websockets/legacy/protocol.py", line 920, in ensure_open
raise self.connection_closed_exc()
websockets.exceptions.ConnectionClosedError: no close frame received or sent`
Because of timeout=-1 the program continues to run as expected once the socket is back. But whenever there an error in wss (ie the above takes place) i want to call a function (to cancel all orders in bybit). How do i call a function on error?
|
closed
|
2021-12-07T16:12:41Z
|
2021-12-11T15:11:28Z
|
https://github.com/bmoscon/cryptofeed/issues/738
|
[
"question"
] |
warproxxx
| 1
|
taverntesting/tavern
|
pytest
| 721
|
How to save a list
|
Hello,
I'm testing a REST API which controls the network configuration of a product.
In the first test stage, I want to backup the current configuration, so that after my actual test stages, I can have one more stage which will restore it.
The problem is that part of this network configuration is stored in lists of unknown length (namely, the IP addresses).
I tried the naive approach:
```yaml
---
test_name: PUT:api/net/:ifname
strict:
- json:off
stages:
- name: Save the configuration of eth0 for further restoration
request:
url: http://{tavern.env_vars.OFORA_HOST}:8080/api/net/eth0
method: GET
response:
status_code: 200
save:
json:
ipv4_method: ipv4.method
ipv6_method: ipv6.method
ipv6_addresses: ipv6.addresses
/* some test stages */
- name: Restore the original configuration
request:
url: http://{tavern.env_vars.OFORA_HOST}:8080/api/net/eth0
json:
autoconnect: False
ipv4:
method: "{ipv4_method:s}"
ipv6:
method: "{ipv6_method:s}"
addresses: "{ipv6_addresses}"
mtu: 0
method: PUT
response:
status_code: 200
```
Which generates the `WARNING tavern.util.dict_util:dict_util.py:46 Formatting 'ipv6_addresses' will result in it being coerced to a string (it is a <class 'box.box_list.BoxList'>)` warning and of course, the API complains that tipe tipe of the value is string when it is expecting an array.
That was apparently to be expected according to the documentation:
> only ‘simple’ values like integers, strings, or float values can be saved. Trying to save a ‘block’ of data such as a JSON list or object is currently unsupported and will cause the test to fail.
I also tried with `$ext` blocks but got the same warning.
Is there a way to save a list and then, retrieve it in order to pass it as a request argument?
Thank you by advance :)
|
closed
|
2021-10-15T08:36:34Z
|
2022-01-04T12:40:45Z
|
https://github.com/taverntesting/tavern/issues/721
|
[] |
ncarrier
| 2
|
OpenBB-finance/OpenBB
|
machine-learning
| 6,825
|
[🕹️] Follow on X
|
### What side quest or challenge are you solving?
Follow on X
### Points
50
### Description
Follow on X
### Provide proof that you've completed the task

|
closed
|
2024-10-20T13:31:20Z
|
2024-10-21T12:58:32Z
|
https://github.com/OpenBB-finance/OpenBB/issues/6825
|
[] |
sateshcharan
| 1
|
ipython/ipython
|
data-science
| 14,799
|
Add typst to the display options
|
[typst](https://typst.app/docs/reference) is gaining serious traction, and rightfully so. I am curios if adding this to the available display types is in the plan: IPython/core/display.py
|
closed
|
2025-02-27T01:17:12Z
|
2025-03-05T12:24:05Z
|
https://github.com/ipython/ipython/issues/14799
|
[] |
aeslaughter
| 3
|
viewflow/viewflow
|
django
| 169
|
'NoneType' object has no attribute '_meta' during ProcessListView rendering
|
Environment:
Request Method: GET
Request URL: http://127.0.0.1:8000/crm/flows/
Django Version: 1.10.5
Python Version: 3.6.0
Installed Applications:
['django.contrib.admin',
'django.contrib.auth',
'django.contrib.contenttypes',
'django.contrib.sessions',
'django.contrib.messages',
'django.contrib.staticfiles',
'django.contrib.humanize',
'material',
'material.frontend',
'material.admin',
'crm.apps.CrmConfig',
'django_mailbox',
'django_markdown',
'viewflow']
Installed Middleware:
['django.middleware.security.SecurityMiddleware',
'django.contrib.sessions.middleware.SessionMiddleware',
'django.middleware.common.CommonMiddleware',
'django.middleware.csrf.CsrfViewMiddleware',
'django.contrib.auth.middleware.AuthenticationMiddleware',
'django.contrib.messages.middleware.MessageMiddleware',
'django.middleware.clickjacking.XFrameOptionsMiddleware']
Traceback:
File "/Users/lukasz/Virtualenvs/miracolo/lib/python3.6/site-packages/django/core/handlers/exception.py" in inner
39. response = get_response(request)
File "/Users/lukasz/Virtualenvs/miracolo/lib/python3.6/site-packages/django/core/handlers/base.py" in _get_response
187. response = self.process_exception_by_middleware(e, request)
File "/Users/lukasz/Virtualenvs/miracolo/lib/python3.6/site-packages/django/core/handlers/base.py" in _get_response
185. response = wrapped_callback(request, *callback_args, **callback_kwargs)
File "/Users/lukasz/Virtualenvs/miracolo/lib/python3.6/site-packages/django/views/generic/base.py" in view
68. return self.dispatch(request, *args, **kwargs)
File "/Users/lukasz/Virtualenvs/miracolo/lib/python3.6/site-packages/django/utils/decorators.py" in _wrapper
67. return bound_func(*args, **kwargs)
File "/Users/lukasz/Virtualenvs/miracolo/lib/python3.6/site-packages/django/contrib/auth/decorators.py" in _wrapped_view
23. return view_func(request, *args, **kwargs)
File "/Users/lukasz/Virtualenvs/miracolo/lib/python3.6/site-packages/django/utils/decorators.py" in bound_func
63. return func.__get__(self, type(self))(*args2, **kwargs2)
File "/Users/lukasz/Virtualenvs/miracolo/lib/python3.6/site-packages/viewflow/views/list.py" in dispatch
216. return super(ProcessListView, self).dispatch(request, *args, **kwargs)
File "/Users/lukasz/Virtualenvs/miracolo/lib/python3.6/site-packages/viewflow/views/base.py" in dispatch
85. super(FlowViewPermissionMixin, self).dispatch)(*args, **kwargs)
File "/Users/lukasz/Virtualenvs/miracolo/lib/python3.6/site-packages/django/contrib/auth/decorators.py" in _wrapped_view
23. return view_func(request, *args, **kwargs)
File "/Users/lukasz/Virtualenvs/miracolo/lib/python3.6/site-packages/django/views/generic/base.py" in dispatch
88. return handler(request, *args, **kwargs)
File "/Users/lukasz/Virtualenvs/miracolo/lib/python3.6/site-packages/django/views/generic/list.py" in get
159. self.object_list = self.get_queryset()
File "/Users/lukasz/Virtualenvs/miracolo/lib/python3.6/site-packages/viewflow/views/list.py" in get_queryset
205. return self.filter.qs
File "/Users/lukasz/Virtualenvs/miracolo/lib/python3.6/site-packages/django_filters/filterset.py" in qs
195. if not self.form.is_valid():
File "/Users/lukasz/Virtualenvs/miracolo/lib/python3.6/site-packages/django_filters/filterset.py" in form
220. for name, filter_ in six.iteritems(self.filters)])
File "/Users/lukasz/Virtualenvs/miracolo/lib/python3.6/site-packages/django_filters/filterset.py" in <listcomp>
220. for name, filter_ in six.iteritems(self.filters)])
File "/Users/lukasz/Virtualenvs/miracolo/lib/python3.6/site-packages/django_filters/filters.py" in field
159. label=self.label, widget=self.widget,
File "/Users/lukasz/Virtualenvs/miracolo/lib/python3.6/site-packages/django_filters/filters.py" in fget
115. model, self.name, self.lookup_expr, self.exclude
File "/Users/lukasz/Virtualenvs/miracolo/lib/python3.6/site-packages/django_filters/utils.py" in label_for_filter
219. name = verbose_field_name(model, field_name)
File "/Users/lukasz/Virtualenvs/miracolo/lib/python3.6/site-packages/django_filters/utils.py" in verbose_field_name
168. parts = get_field_parts(model, field_name)
File "/Users/lukasz/Virtualenvs/miracolo/lib/python3.6/site-packages/django_filters/utils.py" in get_field_parts
86. opts = model._meta
Exception Type: AttributeError at /crm/flows/
Exception Value: 'NoneType' object has no attribute '_meta'
|
closed
|
2017-01-17T12:14:20Z
|
2017-01-20T09:28:47Z
|
https://github.com/viewflow/viewflow/issues/169
|
[] |
lukaszolek
| 2
|
frappe/frappe
|
rest-api
| 31,598
|
minor: css on form-grid error class
|
`develop`
Seems border-radius is incorrect when grid is in error mode ... (no rows on child)

|
closed
|
2025-03-08T21:52:40Z
|
2025-03-24T00:16:46Z
|
https://github.com/frappe/frappe/issues/31598
|
[
"bug"
] |
git-avc
| 0
|
praw-dev/praw
|
api
| 1,817
|
contributor_subreddits returns a list with only subreddits you also moderate, the docs should reflect this
|
**Describe the bug**
Hi folks! According to the docs, `contributor_subreddits()` should `return a ListingGenerator of contributor subreddits. These are subreddits in which the user is an approved user`. According to my testing however, this is only partly true. It does return a list of subreddits in which the authenticated user is a contributor, however the list is restricted to subreddits the user also moderates. Both Reddit (https://www.reddit.com/subreddits/mine/contributor/) and the API (https://www.reddit.com/dev/api#GET_subreddits_mine_contributor) work the same, so it probably works as intended (I assume we can't get the full list from the API). But it wouldn't hurt to add a small notice like such:
> These are subreddits in which the user is **a moderator and** an approved user.
That's just a bit more clear and would help to [prevent confusion](https://www.reddit.com/r/redditdev/comments/oeayi9/how_a_bot_can_know_if_its_approved/). Cheers!
Slightly related to #710 and #246.
**To Reproduce**
```
reddit = praw.Reddit(username=AmputatorBot, ...)
for subreddit in reddit.user.contributor_subreddits(limit=None):
print(str(subreddit))
```
e.g. response:
`AmputatorBot `< moderating (r/AmputatorBot)
`u_AmputatorBot` < self
(..) but missing all non-modded contributing subreddits
**Expected behavior**
The docs say the following:
```
contributor_subreddits(...)
Return a ListingGenerator of contributor subreddits.
These are subreddits in which the user is an approved user.
```
**System Info**
- OS: Windows 10
- Python: 3.7.8
- PRAW Version: 7.4.0
|
closed
|
2021-11-15T19:04:21Z
|
2021-11-16T00:11:04Z
|
https://github.com/praw-dev/praw/issues/1817
|
[] |
jvdburgh
| 4
|
litestar-org/polyfactory
|
pydantic
| 434
|
Bug: Annotations for nested types are ignored
|
### Description
When a pydantic model contains a simple annotated attribute, the restrictions from the annotation are used for generating the value.
Example:
```python
Dimension = Annotated[int, Field(gt=1000, lt=2000)]
class Item(BaseModel):
x: Dimension
class ItemFactory(ModelFactory):
__model__ = Item
print(ItemFactory.build())
```
```
>>> x=1456
```
However, if I change the Item model to
```python
class Item(BaseModel):
dimensions: list[Dimension]
```
I get the following error saying that polyfactory generated the value 8569
```
>>>
pydantic_core._pydantic_core.ValidationError: 1 validation error for Item
dimensions.0
Input should be less than 2000 [type=less_than, input_value=8569, input_type=int]
For further information visit https://errors.pydantic.dev/2.4/v/less_than
```
### URL to code causing the issue
_No response_
### MCVE
```python
from polyfactory.factories.pydantic_factory import ModelFactory
from typing import Annotated
from pydantic import BaseModel, Field
Dimension = Annotated[int, Field(gt=1000, lt=2000)]
class Item(BaseModel):
dimensions: list[Dimension]
class ItemFactory(ModelFactory):
__model__ = Item
print(ItemFactory.build())
```
### Steps to reproduce
```bash
1. Go to '...'
2. Click on '....'
3. Scroll down to '....'
4. See error
```
### Screenshots
"In the format of: ``"
### Logs
_No response_
### Release Version
2.11.0
### Platform
- [X] Linux
- [ ] Mac
- [ ] Windows
- [ ] Other (Please specify in the description above)
|
closed
|
2023-11-07T13:45:19Z
|
2025-03-20T15:53:10Z
|
https://github.com/litestar-org/polyfactory/issues/434
|
[
"bug"
] |
kabell
| 1
|
yunjey/pytorch-tutorial
|
deep-learning
| 197
|
已经解决了win10下的训练自己的数据问题,加Q群857449786 注明pytorch-tutorial 共同研究
|
已经解决了win10下的训练自己的数据问题,加Q群857449786 注明pytorch-tutorial 共同研究
|
open
|
2019-11-23T07:08:16Z
|
2020-08-17T03:31:51Z
|
https://github.com/yunjey/pytorch-tutorial/issues/197
|
[] |
QQ2737499951
| 2
|
influxdata/influxdb-client-python
|
jupyter
| 669
|
Asynchronous client ignoring write precision when set on Point
|
### Specifications
* Client Version: 1.46.0
* InfluxDB Version: 2.7.10
### Code sample to reproduce problem
```python
import asyncio
from influxdb_client import Point, WritePrecision
from influxdb_client.client.influxdb_client_async import InfluxDBClientAsync
_HOST = "http://localhost:8086"
_ORG = "primary"
_TOKEN = "token"
_BUCKET = "MY_BUCKET"
_TIMESTAMP = 1725588000000 # milliseconds, september 6th
async def main() -> None:
p = Point("debug").field("myval", 42).time(_TIMESTAMP, WritePrecision.MS)
client = InfluxDBClientAsync(url=_HOST, token=_TOKEN, org=_ORG)
write_api = client.write_api()
await write_api.write(bucket=_BUCKET, record=p)
await client.close()
if __name__ == "__main__":
asyncio.run(main())
```
### Expected behavior
The write with `InfluxDBClientAsync` to work the same as `InfluxDBClient` does with Point defined write precision.
### Actual behavior
The `InfluxDBClientAsync` does not respect the Point write precision, instead defaults to and uses `DEFAULT_WRITE_PRECISION`.
### Additional info
If the write precision is explicitly stated in the `write_api.write` call, it is handled correctly. Also the docs for the function states that Point data takes precedence which does not happen.
Looking into the calls, I noticed that even though the docstring says that, the `self._serialize` call in the `WriteApiAsync.write` has the `precision_from_point` arguments is set explicitly false. The `self._serialize` call does the extraction right, but the `body` is built incorrectly because it only uses the `write_precision` of the function.
|
closed
|
2024-09-13T13:23:48Z
|
2024-10-09T07:59:57Z
|
https://github.com/influxdata/influxdb-client-python/issues/669
|
[
"bug"
] |
lundaw
| 0
|
microsoft/MMdnn
|
tensorflow
| 467
|
Could not support LeakyReLU and ConvTranspose2d in pytorch to IR
|
Platform (like ubuntu 16.04/win10):win10
Python version:3.5
Source framework with version (like Tensorflow 1.4.1 with GPU):pytorch with GPU
Destination framework with version (like CNTK 2.3 with GPU):caffe
Pre-trained model path (webpath or webdisk path):
Running scripts:mmtoir -f pytorch -d unet --inputShape 1,256,256 -n .\model.pth
|
open
|
2018-10-24T10:25:36Z
|
2018-10-24T10:25:36Z
|
https://github.com/microsoft/MMdnn/issues/467
|
[] |
brisyramshere
| 0
|
gradio-app/gradio
|
data-visualization
| 10,826
|
Tackle iframe resizer issues
|
### Describe the bug
As reported by @jsulz, the presence of the certain components, such as the `gr.Dataframe` can make a Gradio app hosted on Spaces have infinitely long vertical scrolling. We've seen similar issues related to the iframe resizer on other Gradio apps, we should fix this.
@pngwn do you want to take this on?
### Have you searched existing issues? 🔎
- [x] I have searched and found no existing issues
### Reproduction
https://huggingface.co/spaces/huggingface/sizzle/commit/5d4c24bdecc1364c5c4e21af4b6a6cab381264f7 (private Space)
### Screenshot
_No response_
### Logs
```shell
```
### System Info
```shell
gradio==5.20.1
```
### Severity
I can work around it
|
open
|
2025-03-17T21:19:15Z
|
2025-03-20T20:31:49Z
|
https://github.com/gradio-app/gradio/issues/10826
|
[
"bug"
] |
abidlabs
| 1
|
biolab/orange3
|
data-visualization
| 6,175
|
Data Table hangs in in qt6
|
**What's wrong?**
On MacOS with pyqt6 when viewing a large dataset, specifically one with many columns, scrolling horizontally causes Orange to hang.
**How can we reproduce the problem?**
run this in Python Script: (maybe try smaller or larger tables)
```
import numpy as np
from Orange.data import Table, Domain, ContinuousVariable
domain = Domain([ContinuousVariable(str(i)) for i in range(10000)])
arr = np.random.random((100, 10000))
out_data = Table.from_numpy(domain, arr)
```
open in Data Table and drag the scroll bar left and right a bit. (if the widget window is small it tends to handle it a little better)
I assume the problem lies with the item delegates as removing them like so fixed the problem for me.
```
diff --git a/Orange/widgets/data/owtable.py b/Orange/widgets/data/owtable.py
--- a/Orange/widgets/data/owtable.py
+++ b/Orange/widgets/data/owtable.py
@@ -264,7 +264,6 @@ class OWDataTable(OWWidget):
def _create_table_view(self):
view = DataTableView()
view.setSortingEnabled(True)
- view.setItemDelegate(TableDataDelegate(view))
if self.select_rows:
view.setSelectionBehavior(QTableView.SelectRows)
@@ -378,13 +377,6 @@ class OWDataTable(OWWidget):
QColor(*c) for c in data.domain.class_var.colors]
else:
color_schema = None
- if self.show_distributions:
- view.setItemDelegate(
- TableBarItemDelegate(
- view, color=self.dist_color, color_schema=color_schema)
- )
- else:
- view.setItemDelegate(TableDataDelegate(view))
# Enable/disable view sorting based on data's type
view.setSortingEnabled(is_sortable(data))
```
|
closed
|
2022-10-20T09:09:26Z
|
2022-11-29T15:42:21Z
|
https://github.com/biolab/orange3/issues/6175
|
[
"bug"
] |
noahnovsak
| 2
|
streamlit/streamlit
|
deep-learning
| 10,649
|
Preserve `st.column_config.Column` visibility restrictions in the UI
|
### Checklist
- [x] I have searched the [existing issues](https://github.com/streamlit/streamlit/issues) for similar feature requests.
- [x] I added a descriptive title and summary to this issue.
### Summary
Thanks to #10264 we can hide/unhide columns directly from the UI but I have two concerns (one subjective and one that could affect all users) :
1- I expected the "Visibility" icon to always be present in the UI (of course after hovering over the table), even if no columns have been hidden yet (whether via [`st.column_config.Column`](https://docs.streamlit.io/develop/api-reference/data/st.column_config/st.column_config.column) or directly in the UI)

2- In my use case, I load a table from the database (_see code below_) and some columns shouldn’t be exposed to the end user. That’s why I tweak the columns configuration under the hood. But with the new feature, it becomes useless since the column (name + content) can be made visible again from the UI
<details>
<summary>Show code</summary>
```python
import pandas as pd
import streamlit as st
@st.cache_data
def load_data() -> pd.DataFrame:
"""Load the data."""
return pd.DataFrame(
{
"col1": [1, 2, 3],
"col2": [4, 5, 6],
"col3": [7, 8, 9],
"confidential": ["a", "b", "c"],
},
)
st.dataframe(load_data(), column_config={"confidential": {"hidden": True}})
```
</details>

|
open
|
2025-03-05T10:12:27Z
|
2025-03-21T17:06:01Z
|
https://github.com/streamlit/streamlit/issues/10649
|
[
"type:enhancement",
"feature:st.dataframe",
"feature:st.data_editor",
"feature:st.column_config"
] |
VERBOSE-01
| 3
|
automl/auto-sklearn
|
scikit-learn
| 868
|
SIGSEGV
|
auto-sklearn was exactly I was looking for, unfortunately fails even on example from docs:
```python
import sklearn.model_selection
import sklearn.datasets
import sklearn.metrics
import autosklearn.regression
def main():
X, y = sklearn.datasets.load_boston(return_X_y=True)
feature_types = (['numerical'] * 3) + ['categorical'] + (['numerical'] * 9)
X_train, X_test, y_train, y_test = \
sklearn.model_selection.train_test_split(X, y, random_state=1)
automl = autosklearn.regression.AutoSklearnRegressor(
time_left_for_this_task=120,
per_run_time_limit=30,
tmp_folder='/tmp/autosklearn_regression_example_tmp',
output_folder='/tmp/autosklearn_regression_example_out',
)
automl.fit(X_train, y_train, dataset_name='boston',
feat_type=feature_types)
print(automl.show_models())
predictions = automl.predict(X_test)
print("R2 score:", sklearn.metrics.r2_score(y_test, predictions))
if __name__ == '__main__':
main()
```
```
/home/.../.venv/bin/python3.8 /home/.../workspace/varia-private/qtrading/asl.py
/home/.../.venv/lib/python3.8/site-packages/pyparsing.py:3174: FutureWarning: Possible set intersection at position 3
self.re = re.compile(self.reString)
[WARNING] [2020-05-31 16:35:21,635:AutoMLSMBO(1)::boston] Could not find meta-data directory /home/.../.venv/lib/python3.8/site-packages/autosklearn/metalearning/files/r2_regression_dense
Process finished with exit code 139 (interrupted by signal 11: SIGSEGV)
```
OS: Ubuntu 20
Python: 3.8.2
|
closed
|
2020-05-31T14:36:20Z
|
2020-06-09T09:52:20Z
|
https://github.com/automl/auto-sklearn/issues/868
|
[] |
iirekm
| 1
|
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.