repo_name
stringlengths 9
75
| topic
stringclasses 30
values | issue_number
int64 1
203k
| title
stringlengths 1
976
| body
stringlengths 0
254k
| state
stringclasses 2
values | created_at
stringlengths 20
20
| updated_at
stringlengths 20
20
| url
stringlengths 38
105
| labels
listlengths 0
9
| user_login
stringlengths 1
39
| comments_count
int64 0
452
|
|---|---|---|---|---|---|---|---|---|---|---|---|
Gozargah/Marzban
|
api
| 1,086
|
subscription v2ray-json does not support quic
|
Hi, this quic inbound works for v2ray format, but does not work for v2ray-json format, i mean subscription cannot update
inbound:
```
{
"listen": "0.0.0.0",
"port": 3636,
"protocol": "vless",
"settings": {
"clients": [],
"decryption": "none",
"fallbacks": []
},
"sniffing": {
"destOverride": [
"tls",
"quic",
"fakedns"
],
"enabled": true,
"metadataOnly": false,
"routeOnly": false
},
"streamSettings": {
"network": "quic",
"quicSettings": {
"header": {
"type": "none"
},
"key": "key",
"security": "none"
},
"security": "none"
},
"tag": "inbound-3636"
},
```
marzban logs:


|
closed
|
2024-07-04T23:32:48Z
|
2024-08-13T21:35:09Z
|
https://github.com/Gozargah/Marzban/issues/1086
|
[
"Bug"
] |
m0x61h0x64i
| 2
|
mars-project/mars
|
pandas
| 2,585
|
[BUG] TimeoutError: Timeout in request queue
|
When fetching dataframe to local, when chunks is greater than 200, following errors happen:
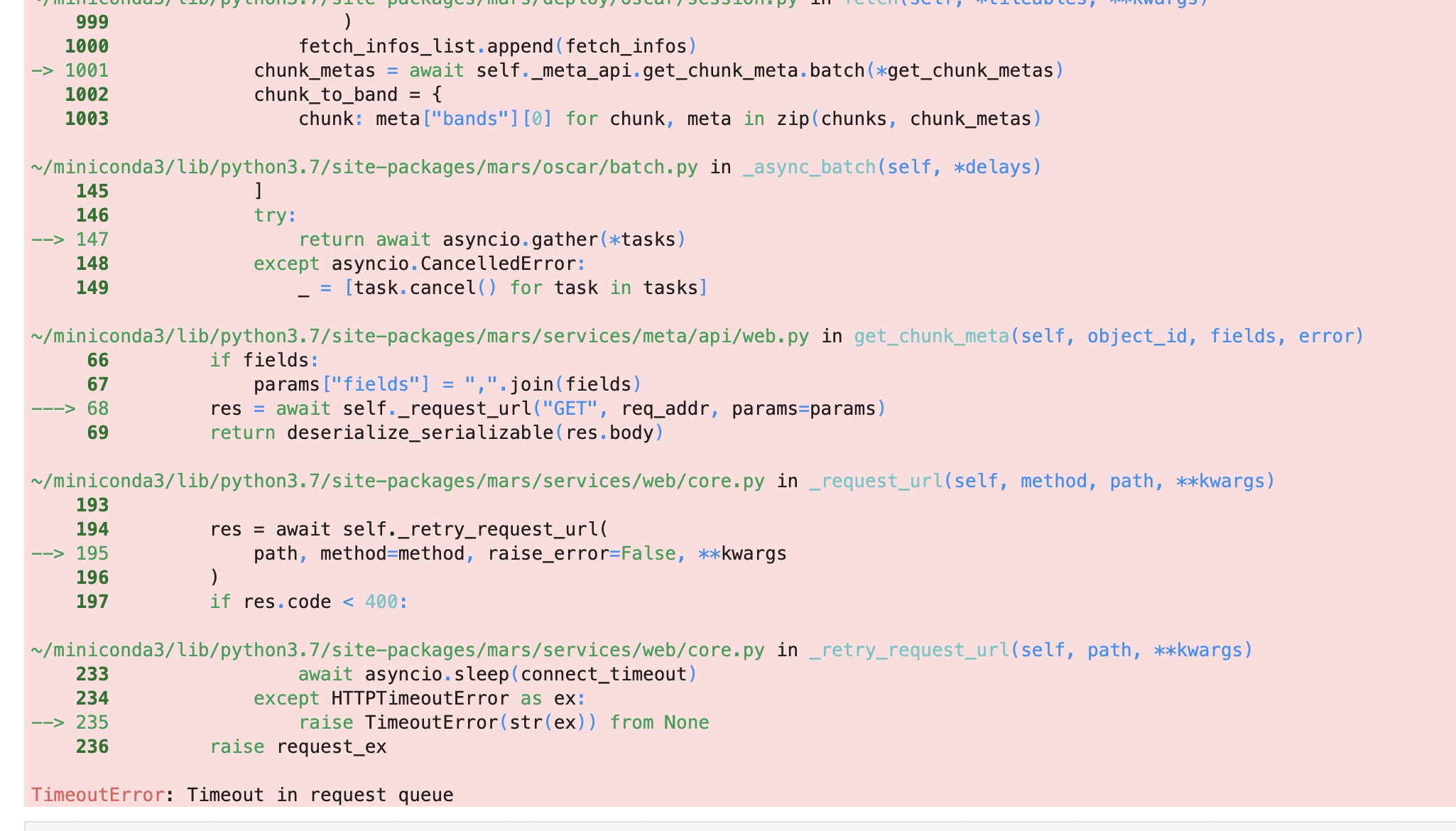
|
open
|
2021-11-25T10:35:52Z
|
2021-11-25T10:35:52Z
|
https://github.com/mars-project/mars/issues/2585
|
[] |
chaokunyang
| 0
|
roboflow/supervision
|
machine-learning
| 1,694
|
Crash when filtering empty detections: xyxy shape (0, 0, 4).
|
Reproduction code:
```python
import supervision as sv
import numpy as np
CLASSES = [0, 1, 2]
prediction = sv.Detections.empty()
prediction = prediction[np.isin(prediction["class_name"], CLASSES)]
```
Error:
```
Traceback (most recent call last):
File "/Users/linasko/.settler_workspace/pr/supervision-fresh/run_detections.py", line 7, in <module>
prediction = prediction[np.isin(prediction["class_name"], CLASSES)]
File "/Users/linasko/.settler_workspace/pr/supervision-fresh/supervision/detection/core.py", line 1206, in __getitem__
return Detections(
File "<string>", line 10, in __init__
File "/Users/linasko/.settler_workspace/pr/supervision-fresh/supervision/detection/core.py", line 144, in __post_init__
validate_detections_fields(
File "/Users/linasko/.settler_workspace/pr/supervision-fresh/supervision/validators/__init__.py", line 120, in validate_detections_fields
validate_xyxy(xyxy)
File "/Users/linasko/.settler_workspace/pr/supervision-fresh/supervision/validators/__init__.py", line 11, in validate_xyxy
raise ValueError(
ValueError: xyxy must be a 2D np.ndarray with shape (_, 4), but got shape (0, 0, 4)
```
|
closed
|
2024-11-28T11:31:18Z
|
2024-12-04T10:15:33Z
|
https://github.com/roboflow/supervision/issues/1694
|
[
"bug"
] |
LinasKo
| 0
|
flasgger/flasgger
|
rest-api
| 146
|
OpenAPI 3.0
|
https://www.youtube.com/watch?v=wBDSR0x3GZo
|
open
|
2017-08-10T17:42:13Z
|
2020-07-16T10:23:14Z
|
https://github.com/flasgger/flasgger/issues/146
|
[
"hacktoberfest"
] |
rochacbruno
| 10
|
piskvorky/gensim
|
machine-learning
| 3,536
|
scipy probably not needed in [build-system.requires] table
|
<!--
**IMPORTANT**:
- Use the [Gensim mailing list](https://groups.google.com/g/gensim) to ask general or usage questions. Github issues are only for bug reports.
- Check [Recipes&FAQ](https://github.com/RaRe-Technologies/gensim/wiki/Recipes-&-FAQ) first for common answers.
Github bug reports that do not include relevant information and context will be closed without an answer. Thanks!
-->
#### Problem description
I believe that specifying `scipy` as a build-only dependency is unnecessary.
Pip builds the library in isolated environment (by default) where it first downloads (and alternatively builds) build-only dependencies. This behaviour creates additional problems for architectures for which there are not many _.whl_ distributions, like e.g. **ppc64le**.
#### Steps/code/corpus to reproduce
1) create an environment where there is already an older `scipy` library version installed
2)
```sh
cd gensim/
pip install .
```
3) As there is no `gensim` _.whl_ distribution for **ppc64le**, pip will try building `gensim` in a new isolated environment with build-only dependencies.
As there is no `scipy` _.whl_ distribution for ppc64le either, pip will try building that as well.
Despite the fact that `scipy` is already installed in desired version within the target environment.
Not only there will be a `scipy` version mismatch (pip will be building the latest version in the isolated environment) but it will also:
* significantly prolong the install phase, as `scipy` takes relatively long to build from source,
* create additional dependencies mess, as `scipy` build requires multiple other dependencies and system-level libraries
#### Desired resolution
I've tested locally installing gensim with modified `pyproject.toml` file (deleted `scipy`) and it works as expected.
Is there any other logic that does not allow deleting `scipy` as a build-only dependency?
#### Versions
`gensim>4.3.*`
|
closed
|
2024-06-06T13:42:33Z
|
2024-07-18T12:03:09Z
|
https://github.com/piskvorky/gensim/issues/3536
|
[
"awaiting reply"
] |
filip-komarzyniec
| 2
|
albumentations-team/albumentations
|
deep-learning
| 2,097
|
[Add transform] Add RandomJPEG
|
Add RandomJPEG which is a child of ImageCompression and has the same API as Kornia's
https://kornia.readthedocs.io/en/latest/augmentation.module.html#kornia.augmentation.RandomJPEG
|
closed
|
2024-11-08T15:50:40Z
|
2024-11-09T00:58:42Z
|
https://github.com/albumentations-team/albumentations/issues/2097
|
[
"enhancement"
] |
ternaus
| 0
|
gyli/PyWaffle
|
data-visualization
| 2
|
width problems with a thousand blocks
|
When plotting a larger number of blocks, the width of the white space between them become unstable:
```
plt.figure(
FigureClass=Waffle,
rows=20,
columns=80,
values=[300, 700],
figsize=(18, 10)
);
plt.savefig('example.png')
```

This is probably outside the original scope of the package and maybe should even be discouraged, but sometimes is useful to give the reader the impression of dealing with a large population. Feel free to close this issue.
|
open
|
2017-11-23T17:27:12Z
|
2019-10-06T22:29:14Z
|
https://github.com/gyli/PyWaffle/issues/2
|
[] |
lincolnfrias
| 2
|
pyjanitor-devs/pyjanitor
|
pandas
| 1,200
|
[BUG] `deprecated_kwargs` (list[str]) in v0.24 raises type object not subscriptable error
|
# Brief Description
The addition of deprecated_kwargs in version 0.23 causes a type object not subscriptable error.
# System Information
I'm using Python 3.8.12 on a sagemaker instance. I'm pretty sure this is the issue, that my company has us locked at 3.8.12 right now. Selecting the v.0.23 does solve the problem.
I'm sorry if this isn't enough information at the moment, let me know if you need anything else.
# Error
```
TypeError Traceback (most recent call last)
/tmp/ipykernel_18038/2902872131.py in <cell line: 1>()
----> 1 import janitor
~/anaconda3/envs/python3/lib/python3.8/site-packages/janitor/__init__.py in <module>
7
8 from .accessors import * # noqa: F403, F401
----> 9 from .functions import * # noqa: F403, F401
10 from .io import * # noqa: F403, F401
11 from .math import * # noqa: F403, F401
~/anaconda3/envs/python3/lib/python3.8/site-packages/janitor/functions/__init__.py in <module>
17
18
---> 19 from .add_columns import add_columns
20 from .also import also
21 from .bin_numeric import bin_numeric
~/anaconda3/envs/python3/lib/python3.8/site-packages/janitor/functions/add_columns.py in <module>
1 import pandas_flavor as pf
2
----> 3 from janitor.utils import check, deprecated_alias
4 import pandas as pd
5 from typing import Union, List, Any, Tuple
~/anaconda3/envs/python3/lib/python3.8/site-packages/janitor/utils.py in <module>
214
215 def deprecated_kwargs(
--> 216 *arguments: list[str],
217 message: str = (
218 "The keyword argument '{argument}' of '{func_name}' is deprecated."
TypeError: 'type' object is not subscriptable
```
|
closed
|
2022-11-14T16:11:14Z
|
2022-11-21T06:04:41Z
|
https://github.com/pyjanitor-devs/pyjanitor/issues/1200
|
[
"bug"
] |
zykezero
| 11
|
modelscope/modelscope
|
nlp
| 1,122
|
from modelscope.msdatasets import MsDataset 报错
|
(Pdb) from modelscope.msdatasets import MsDataset
*** ModuleNotFoundError: No module named 'datasets.download'
(Pdb) import modelscope
(Pdb) modelscope.__version__
'1.17.0'
(Pdb) datasets.__version__
'2.0.0'
Python 3.10.15,ubuntu 22.04 系统
当前modescope 需要使用哪个版本的datasets ?
|
closed
|
2024-12-04T10:15:02Z
|
2024-12-19T12:13:28Z
|
https://github.com/modelscope/modelscope/issues/1122
|
[] |
robator0127
| 1
|
microsoft/UFO
|
automation
| 190
|
Batch Mode and Follower Mode get "No module named 'ufo.config'; 'ufo' is not a package" exception
|
When trying the steps with [Batch Mode](https://microsoft.github.io/UFO/advanced_usage/batch_mode/) and [Follower Mode](https://microsoft.github.io/UFO/advanced_usage/follower_mode/) based on the document, it will throw "ModuleNotFoundError: No module named 'ufo.config'; 'ufo' is not a package" exception which result to the command cannot be executed.
**Here is the repro steps:**
Assume the Plan file is prepared based on the document.
1. Open Command Prompt Window and navigate to the cloned UFO folder.
2. Run "python ufo\ufo.py --task_name testbatchmode --mode batch_normal --plan "parentpath\planfilename.json"".
**Expected Result:**
Command will be run without errors.
**Actually Result:**
Failed with below error:
```
Traceback (most recent call last):
File "E:\Repos\UFO\ufo\ufo.py", line 7, in <module>
from ufo.config.config import Config
File "E:\Repos\UFO\ufo\ufo.py", line 7, in <module>
from ufo.config.config import Config
ModuleNotFoundError: No module named 'ufo.config'; 'ufo' is not a package
```
**What we have did:**
We tried use pip install command to install ufo or ufo.config, but both them are could not be found:

We also tried with the newest vyokky/dev branch, but the error still exists.
|
open
|
2025-03-19T06:33:06Z
|
2025-03-19T08:28:24Z
|
https://github.com/microsoft/UFO/issues/190
|
[] |
WeiweiCaiAcpt
| 2
|
automl/auto-sklearn
|
scikit-learn
| 1,573
|
Add pylint linter
|
After we have removed all mypy ignores.
|
open
|
2022-08-22T11:23:17Z
|
2022-08-24T04:04:50Z
|
https://github.com/automl/auto-sklearn/issues/1573
|
[
"maintenance"
] |
mfeurer
| 0
|
RobertCraigie/prisma-client-py
|
pydantic
| 106
|
Experimental support for the Decimal type
|
## Why is this experimental?
Currently Prisma Client Python does not have access to the field metadata containing the precision of `Decimal` fields at the database level. This means that we cannot:
- Raise an error if you attempt to pass a `Decimal` value with a greater precision than the database supports, leading to implicit truncation which may cause confusing errors
- Set the precision level on the returned `decimal.Decimal` objects to match the database level, potentially leading to even more confusing errors.
To try and mitigate the effects of these errors you must be explicit that you understand that the support for the `Decimal` type is not up to the standard of the other types. You do this by setting the `enable_experimental_decimal` config flag, e.g.
```prisma
generator py {
provider = "prisma-client-py"
enable_experimental_decimal = true
}
```
|
closed
|
2021-11-07T23:58:44Z
|
2022-03-24T22:03:06Z
|
https://github.com/RobertCraigie/prisma-client-py/issues/106
|
[
"topic: types",
"kind/feature",
"level/advanced",
"priority/medium"
] |
RobertCraigie
| 12
|
graphdeco-inria/gaussian-splatting
|
computer-vision
| 986
|
Error when installing the SIBR viewer on Ubuntu 22.04
|
Hi! I had this error when I ran the installation command
`cmake -Bbuild . -DCMAKE_BUILD_TYPE=Release`
> There is no provided OpenCV library for your compiler, relying on find_package to find it
-- Found OpenCV: /usr (found suitable version "4.5.4", minimum required is "4.5")
-- Populating library imgui...
-- Populating library nativefiledialog...
-- Checking for module 'gtk+-3.0'
-- Package 'Lerc', required by 'libtiff-4', not found
CMake Error at /usr/share/cmake-3.22/Modules/FindPkgConfig.cmake:603 (message):
A required package was not found
Call Stack (most recent call first):
/usr/share/cmake-3.22/Modules/FindPkgConfig.cmake:825 (_pkg_check_modules_internal)
extlibs/nativefiledialog/nativefiledialog/CMakeLists.txt:20 (pkg_check_modules)
-- Configuring incomplete, errors occurred!
See also "/home/yiduo/projects/code/gs/SIBR_viewers/build/CMakeFiles/CMakeOutput.log".
System: Ubuntu 22.04.5 LTS
Cmake version: 3.22.1

It seems like this is not necessarily related to the codebase here. But does anyone have any idea how to solve this? Thanks a lot!
|
open
|
2024-09-13T00:09:47Z
|
2024-09-13T00:09:47Z
|
https://github.com/graphdeco-inria/gaussian-splatting/issues/986
|
[] |
yiduohao
| 0
|
koxudaxi/datamodel-code-generator
|
fastapi
| 1,982
|
AttributeError: 'FieldInfo' object has no attribute '<EnumName>'
|
**Describe the bug**
Generating from a schema with an Enum type causes `AttributeError: 'FieldInfo' object has no attribute '<EnumName>'`
**To Reproduce**
File structure after codegen should look like:
```
schemas/
├─ bean.json
├─ bean_type.json
src/
├─ __init__.py
├─ bean.py
├─ bean_type.py
main.py
```
With the schemas defined as follows:
`schemas/bean.json`
```json
{
"$schema": "https://json-schema.org/draft/2020-12/schema",
"$id": "bean.json",
"type": "object",
"title": "Bean",
"properties": {
"beanType": { "$ref": "bean_type.json" },
"name": { "type": "string" }
},
"additionalProperties": false,
"required": ["beanType", "name"]
}
```
`schemas/bean_type.json`
```json
{
"$schema": "https://json-schema.org/draft/2020-12/schema",
"$id": "bean_type.json",
"title": "BeanType",
"additionalProperties": false,
"enum": ["STRING_BEAN", "RUNNER_BEAN", "GREEN_BEAN", "BAKED_BEAN"]
}
```
and
`main.py`
```py
from src.bean import Bean
if __name__ == "__main__":
pass
```
**Used commandline**
```
$ datamodel-codegen \
--use-title-as-name \
--use-standard-collections \
--snake-case-field \
--target-python-version 3.12 \
--input schemas \
--input-file-type jsonschema \
--output src
```
**Expected behavior**
Exactly what happened, except we should then be able to import and use the generated classes. Instead, an AttributeError is raised.
**Version:**
- OS: MacOS 14.3.1 (23D60)
- Python version: 3.12.3
- Pydantic version: 2.7.2
- datamodel-code-generator version: 0.25.6
**Additional context**
In the generated file `src/bean.py`, if we manually change `from . import bean_type` to `from .bean_type import BeanType`, and the corresponding usage in the `Bean` class definition, the error disappears. Might be related to #1683 / #1684
|
closed
|
2024-06-02T15:01:30Z
|
2024-06-18T05:14:07Z
|
https://github.com/koxudaxi/datamodel-code-generator/issues/1982
|
[] |
alpoi-x
| 0
|
tensorflow/tensor2tensor
|
machine-learning
| 1,523
|
The evolved transformer code is the final graph or the whole procedure to find the best graph?
|
I'm new to neural architecture search. Thank you.
|
open
|
2019-03-25T02:27:41Z
|
2020-11-12T15:56:57Z
|
https://github.com/tensorflow/tensor2tensor/issues/1523
|
[] |
guotong1988
| 6
|
ghtmtt/DataPlotly
|
plotly
| 257
|
Display every record as a line in a scatter plot
|
Hi,
**Short Feature Explanation**
I am wondering if it would be possible to create a scatter plot that displays a line per record instead of a point per record.
This would be done by selecting two columns storing arrays of values for the x and y fields.
For example, with a table: Temp(xs int[], ys[]), selecting the xs and ys columns for the x and y fields respectively would create a scatterplot with as many lines as records in table A, and as many points per line as values in the xs and ys arrays. (Of course, xs and ys should have the same amount of elements each)
**Context**
To explain my problem, I am a developer of [MobilityDB](https://github.com/MobilityDB/MobilityDB), and I am trying to display temporal properties in QGIS using DataPlotly. An example of a table that we want to display would be:
Ports(name text, port geometry(Polygon), shipsInside tint)
Every record thus represents a port and has an attribute storing the number of ships inside this port over time. The ports are represented as polygons on the map, and I would thus like to represent the 'shipsInside' attribute as a line on a scatter plot.
For simplicity, let's assume that this temporal attribute is stored in two columns: one containing an array of timestamps, and one containing an array of values: (this can be done in practice as well)
Ports(name text, port geometry, ts timestamptz[], vals int[])
**Current Workaround**
Currently, I can display a single record of the original table by creating a new table for it:
Ports_temp(name text, port geometry, t timestamptz, val int)
This table contains a record for each pair of (t, val) in the arrays of the original record.
Using this table, I can then create a scatterplot using t and val as the x and y fields respectively.
Of course, this solution is not ideal, since this demands a new table for each record of the original Ports table.
**Conclusion**
I am thus wondering how hard it would be to allow scatterplots to display such records with temporal attributes as lines on a scatterplot.
Ideally, this should be done either by selecting two columns that store arrays of values or selecting a single temporal column (tint, float or tbool).
Best Regards,
Maxime Schoemans
|
open
|
2021-03-15T17:03:19Z
|
2021-03-24T12:58:10Z
|
https://github.com/ghtmtt/DataPlotly/issues/257
|
[
"enhancement"
] |
mschoema
| 6
|
apache/airflow
|
python
| 47,970
|
"consuming_dags" and "producing_tasks" do not correct account for Asset.ref
|
### Body
They are direct SQLAlchemy relationships to only concrete references (DagAssetScheduleReference and TaskOutletAssetReference). Not quite sure how to fix this. Maybe they should be plain properties that return list-of-union instead? We don’t really need those relationships….
### Committer
- [x] I acknowledge that I am a maintainer/committer of the Apache Airflow project.
|
open
|
2025-03-19T18:23:11Z
|
2025-03-19T18:29:31Z
|
https://github.com/apache/airflow/issues/47970
|
[
"kind:bug",
"area:datasets"
] |
uranusjr
| 1
|
coqui-ai/TTS
|
deep-learning
| 3,177
|
[Bug] Loading XTTS via Xtts.load_checkpoint()
|
### Describe the bug
When loading the model using `Xtts.load_checkpoint`, exception is raised as `Error(s) in loading state_dict for Xtts`, which leads to missing keys GPT embedding weights and size mismatch on Mel embedding. Even tried providing the directory which had base(v2) model checkpoints and got the same result.
### To Reproduce
```
import os
import torch
import torchaudio
from TTS.tts.configs.xtts_config import XttsConfig
from TTS.tts.models.xtts import Xtts
print("Loading model...")
config = XttsConfig()
config.load_json("/path/to/xtts/config.json")
model = Xtts.init_from_config(config)
model.load_checkpoint(config, checkpoint_dir="/path/to/xtts/", use_deepspeed=True)
model.cuda()
print("Computing speaker latents...")
gpt_cond_latent, speaker_embedding = model.get_conditioning_latents(audio_path=["reference.wav"])
print("Inference...")
out = model.inference(
"It took me quite a long time to develop a voice and now that I have it I am not going to be silent.",
"en",
gpt_cond_latent,
speaker_embedding,
temperature=0.7, # Add custom parameters here
)
torchaudio.save("xtts.wav", torch.tensor(out["wav"]).unsqueeze(0), 24000)
```
### Expected behavior
Load the checkpoint and run inference without exception.
### Logs
```shell
11-08 22:13:53 [__main__ ] ERROR - Error(s) in loading state_dict for Xtts:
Missing key(s) in state_dict: "gpt.gpt.wte.weight", "gpt.prompt_embedding.weight", "gpt.prompt_pos_embedding.emb.weight", "gpt.gpt_inference.transformer.h.0.ln_1.weight", "gpt.gpt_inference.transformer.h.0.ln_1.bias", "gpt.gpt_inference.transformer.h.0.attn.c_attn.weight", "gpt.gpt_inference.transformer.h.0.attn.c_attn.bias", "gpt.gpt_inference.transformer.h.0.attn.c_proj.weight", "gpt.gpt_inference.transformer.h.0.attn.c_proj.bias", "gpt.gpt_inference.transformer.h.0.ln_2.weight", "gpt.gpt_inference.transformer.h.0.ln_2.bias", "gpt.gpt_inference.transformer.h.0.mlp.c_fc.weight", "gpt.gpt_inference.transformer.h.0.mlp.c_fc.bias", "gpt.gpt_inference.transformer.h.0.mlp.c_proj.weight", "gpt.gpt_inference.transformer.h.0.mlp.c_proj.bias", "gpt.gpt_inference.transformer.h.1.ln_1.weight", "gpt.gpt_inference.transformer.h.1.ln_1.bias", "gpt.gpt_inference.transformer.h.1.attn.c_attn.weight", "gpt.gpt_inference.transformer.h.1.attn.c_attn.bias", "gpt.gpt_inference.transformer.h.1.attn.c_proj.weight", "gpt.gpt_inference.transformer.h.1.attn.c_proj.bias", "gpt.gpt_inference.transformer.h.1.ln_2.weight", "gpt.gpt_inference.transformer.h.1.ln_2.bias", "gpt.gpt_inference.transformer.h.1.mlp.c_fc.weight", "gpt.gpt_inference.transformer.h.1.mlp.c_fc.bias", "gpt.gpt_inference.transformer.h.1.mlp.c_proj.weight", "gpt.gpt_inference.transformer.h.1.mlp.c_proj.bias", "gpt.gpt_inference.transformer.h.2.ln_1.weight", "gpt.gpt_inference.transformer.h.2.ln_1.bias", "gpt.gpt_inference.transformer.h.2.attn.c_attn.weight", "gpt.gpt_inference.transformer.h.2.attn.c_attn.bias", "gpt.gpt_inference.transformer.h.2.attn.c_proj.weight", "gpt.gpt_inference.transformer.h.2.attn.c_proj.bias", "gpt.gpt_inference.transformer.h.2.ln_2.weight", "gpt.gpt_inference.transformer.h.2.ln_2.bias", "gpt.gpt_inference.transformer.h.2.mlp.c_fc.weight", "gpt.gpt_inference.transformer.h.2.mlp.c_fc.bias", "gpt.gpt_inference.transformer.h.2.mlp.c_proj.weight", "gpt.gpt_inference.transformer.h.2.mlp.c_proj.bias", "gpt.gpt_inference.transformer.h.3.ln_1.weight", "gpt.gpt_inference.transformer.h.3.ln_1.bias", "gpt.gpt_inference.transformer.h.3.attn.c_attn.weight", "gpt.gpt_inference.transformer.h.3.attn.c_attn.bias", "gpt.gpt_inference.transformer.h.3.attn.c_proj.weight", "gpt.gpt_inference.transformer.h.3.attn.c_proj.bias", "gpt.gpt_inference.transformer.h.3.ln_2.weight", "gpt.gpt_inference.transformer.h.3.ln_2.bias", "gpt.gpt_inference.transformer.h.3.mlp.c_fc.weight", "gpt.gpt_inference.transformer.h.3.mlp.c_fc.bias", "gpt.gpt_inference.transformer.h.3.mlp.c_proj.weight", "gpt.gpt_inference.transformer.h.3.mlp.c_proj.bias", "gpt.gpt_inference.transformer.h.4.ln_1.weight", "gpt.gpt_inference.transformer.h.4.ln_1.bias", "gpt.gpt_inference.transformer.h.4.attn.c_attn.weight", "gpt.gpt_inference.transformer.h.4.attn.c_attn.bias", "gpt.gpt_inference.transformer.h.4.attn.c_proj.weight", "gpt.gpt_inference.transformer.h.4.attn.c_proj.bias", "gpt.gpt_inference.transformer.h.4.ln_2.weight", "gpt.gpt_inference.transformer.h.4.ln_2.bias", "gpt.gpt_inference.transformer.h.4.mlp.c_fc.weight", "gpt.gpt_inference.transformer.h.4.mlp.c_fc.bias", "gpt.gpt_inference.transformer.h.4.mlp.c_proj.weight", "gpt.gpt_inference.transformer.h.4.mlp.c_proj.bias", "gpt.gpt_inference.transformer.h.5.ln_1.weight", "gpt.gpt_inference.transformer.h.5.ln_1.bias", "gpt.gpt_inference.transformer.h.5.attn.c_attn.weight", "gpt.gpt_inference.transformer.h.5.attn.c_attn.bias", "gpt.gpt_inference.transformer.h.5.attn.c_proj.weight", "gpt.gpt_inference.transformer.h.5.attn.c_proj.bias", "gpt.gpt_inference.transformer.h.5.ln_2.weight", "gpt.gpt_inference.transformer.h.5.ln_2.bias", "gpt.gpt_inference.transformer.h.5.mlp.c_fc.weight", "gpt.gpt_inference.transformer.h.5.mlp.c_fc.bias", "gpt.gpt_inference.transformer.h.5.mlp.c_proj.weight", "gpt.gpt_inference.transformer.h.5.mlp.c_proj.bias", "gpt.gpt_inference.transformer.h.6.ln_1.weight", "gpt.gpt_inference.transformer.h.6.ln_1.bias", "gpt.gpt_inference.transformer.h.6.attn.c_attn.weight", "gpt.gpt_inference.transformer.h.6.attn.c_attn.bias", "gpt.gpt_inference.transformer.h.6.attn.c_proj.weight", "gpt.gpt_inference.transformer.h.6.attn.c_proj.bias", "gpt.gpt_inference.transformer.h.6.ln_2.weight", "gpt.gpt_inference.transformer.h.6.ln_2.bias", "gpt.gpt_inference.transformer.h.6.mlp.c_fc.weight", "gpt.gpt_inference.transformer.h.6.mlp.c_fc.bias", "gpt.gpt_inference.transformer.h.6.mlp.c_proj.weight", "gpt.gpt_inference.transformer.h.6.mlp.c_proj.bias", "gpt.gpt_inference.transformer.h.7.ln_1.weight", "gpt.gpt_inference.transformer.h.7.ln_1.bias", "gpt.gpt_inference.transformer.h.7.attn.c_attn.weight", "gpt.gpt_inference.transformer.h.7.attn.c_attn.bias", "gpt.gpt_inference.transformer.h.7.attn.c_proj.weight", "gpt.gpt_inference.transformer.h.7.attn.c_proj.bias", "gpt.gpt_inference.transformer.h.7.ln_2.weight", "gpt.gpt_inference.transformer.h.7.ln_2.bias", "gpt.gpt_inference.transformer.h.7.mlp.c_fc.weight", "gpt.gpt_inference.transformer.h.7.mlp.c_fc.bias", "gpt.gpt_inference.transformer.h.7.mlp.c_proj.weight", "gpt.gpt_inference.transformer.h.7.mlp.c_proj.bias", "gpt.gpt_inference.transformer.h.8.ln_1.weight", "gpt.gpt_inference.transformer.h.8.ln_1.bias", "gpt.gpt_inference.transformer.h.8.attn.c_attn.weight", "gpt.gpt_inference.transformer.h.8.attn.c_attn.bias", "gpt.gpt_inference.transformer.h.8.attn.c_proj.weight", "gpt.gpt_inference.transformer.h.8.attn.c_proj.bias", "gpt.gpt_inference.transformer.h.8.ln_2.weight", "gpt.gpt_inference.transformer.h.8.ln_2.bias", "gpt.gpt_inference.transformer.h.8.mlp.c_fc.weight", "gpt.gpt_inference.transformer.h.8.mlp.c_fc.bias", "gpt.gpt_inference.transformer.h.8.mlp.c_proj.weight", "gpt.gpt_inference.transformer.h.8.mlp.c_proj.bias", "gpt.gpt_inference.transformer.h.9.ln_1.weight", "gpt.gpt_inference.transformer.h.9.ln_1.bias", "gpt.gpt_inference.transformer.h.9.attn.c_attn.weight", "gpt.gpt_inference.transformer.h.9.attn.c_attn.bias", "gpt.gpt_inference.transformer.h.9.attn.c_proj.weight", "gpt.gpt_inference.transformer.h.9.attn.c_proj.bias", "gpt.gpt_inference.transformer.h.9.ln_2.weight", "gpt.gpt_inference.transformer.h.9.ln_2.bias", "gpt.gpt_inference.transformer.h.9.mlp.c_fc.weight", "gpt.gpt_inference.transformer.h.9.mlp.c_fc.bias", "gpt.gpt_inference.transformer.h.9.mlp.c_proj.weight", "gpt.gpt_inference.transformer.h.9.mlp.c_proj.bias", "gpt.gpt_inference.transformer.h.10.ln_1.weight", "gpt.gpt_inference.transformer.h.10.ln_1.bias", "gpt.gpt_inference.transformer.h.10.attn.c_attn.weight", "gpt.gpt_inference.transformer.h.10.attn.c_attn.bias", "gpt.gpt_inference.transformer.h.10.attn.c_proj.weight", "gpt.gpt_inference.transformer.h.10.attn.c_proj.bias", "gpt.gpt_inference.transformer.h.10.ln_2.weight", "gpt.gpt_inference.transformer.h.10.ln_2.bias", "gpt.gpt_inference.transformer.h.10.mlp.c_fc.weight", "gpt.gpt_inference.transformer.h.10.mlp.c_fc.bias", "gpt.gpt_inference.transformer.h.10.mlp.c_proj.weight", "gpt.gpt_inference.transformer.h.10.mlp.c_proj.bias", "gpt.gpt_inference.transformer.h.11.ln_1.weight", "gpt.gpt_inference.transformer.h.11.ln_1.bias", "gpt.gpt_inference.transformer.h.11.attn.c_attn.weight", "gpt.gpt_inference.transformer.h.11.attn.c_attn.bias", "gpt.gpt_inference.transformer.h.11.attn.c_proj.weight", "gpt.gpt_inference.transformer.h.11.attn.c_proj.bias", "gpt.gpt_inference.transformer.h.11.ln_2.weight", "gpt.gpt_inference.transformer.h.11.ln_2.bias", "gpt.gpt_inference.transformer.h.11.mlp.c_fc.weight", "gpt.gpt_inference.transformer.h.11.mlp.c_fc.bias", "gpt.gpt_inference.transformer.h.11.mlp.c_proj.weight", "gpt.gpt_inference.transformer.h.11.mlp.c_proj.bias", "gpt.gpt_inference.transformer.h.12.ln_1.weight", "gpt.gpt_inference.transformer.h.12.ln_1.bias", "gpt.gpt_inference.transformer.h.12.attn.c_attn.weight", "gpt.gpt_inference.transformer.h.12.attn.c_attn.bias", "gpt.gpt_inference.transformer.h.12.attn.c_proj.weight", "gpt.gpt_inference.transformer.h.12.attn.c_proj.bias", "gpt.gpt_inference.transformer.h.12.ln_2.weight", "gpt.gpt_inference.transformer.h.12.ln_2.bias", "gpt.gpt_inference.transformer.h.12.mlp.c_fc.weight", "gpt.gpt_inference.transformer.h.12.mlp.c_fc.bias", "gpt.gpt_inference.transformer.h.12.mlp.c_proj.weight", "gpt.gpt_inference.transformer.h.12.mlp.c_proj.bias", "gpt.gpt_inference.transformer.h.13.ln_1.weight", "gpt.gpt_inference.transformer.h.13.ln_1.bias", "gpt.gpt_inference.transformer.h.13.attn.c_attn.weight", "gpt.gpt_inference.transformer.h.13.attn.c_attn.bias", "gpt.gpt_inference.transformer.h.13.attn.c_proj.weight", "gpt.gpt_inference.transformer.h.13.attn.c_proj.bias", "gpt.gpt_inference.transformer.h.13.ln_2.weight", "gpt.gpt_inference.transformer.h.13.ln_2.bias", "gpt.gpt_inference.transformer.h.13.mlp.c_fc.weight", "gpt.gpt_inference.transformer.h.13.mlp.c_fc.bias", "gpt.gpt_inference.transformer.h.13.mlp.c_proj.weight", "gpt.gpt_inference.transformer.h.13.mlp.c_proj.bias", "gpt.gpt_inference.transformer.h.14.ln_1.weight", "gpt.gpt_inference.transformer.h.14.ln_1.bias", "gpt.gpt_inference.transformer.h.14.attn.c_attn.weight", "gpt.gpt_inference.transformer.h.14.attn.c_attn.bias", "gpt.gpt_inference.transformer.h.14.attn.c_proj.weight", "gpt.gpt_inference.transformer.h.14.attn.c_proj.bias", "gpt.gpt_inference.transformer.h.14.ln_2.weight", "gpt.gpt_inference.transformer.h.14.ln_2.bias", "gpt.gpt_inference.transformer.h.14.mlp.c_fc.weight", "gpt.gpt_inference.transformer.h.14.mlp.c_fc.bias", "gpt.gpt_inference.transformer.h.14.mlp.c_proj.weight", "gpt.gpt_inference.transformer.h.14.mlp.c_proj.bias", "gpt.gpt_inference.transformer.h.15.ln_1.weight", "gpt.gpt_inference.transformer.h.15.ln_1.bias", "gpt.gpt_inference.transformer.h.15.attn.c_attn.weight", "gpt.gpt_inference.transformer.h.15.attn.c_attn.bias", "gpt.gpt_inference.transformer.h.15.attn.c_proj.weight", "gpt.gpt_inference.transformer.h.15.attn.c_proj.bias", "gpt.gpt_inference.transformer.h.15.ln_2.weight", "gpt.gpt_inference.transformer.h.15.ln_2.bias", "gpt.gpt_inference.transformer.h.15.mlp.c_fc.weight", "gpt.gpt_inference.transformer.h.15.mlp.c_fc.bias", "gpt.gpt_inference.transformer.h.15.mlp.c_proj.weight", "gpt.gpt_inference.transformer.h.15.mlp.c_proj.bias", "gpt.gpt_inference.transformer.h.16.ln_1.weight", "gpt.gpt_inference.transformer.h.16.ln_1.bias", "gpt.gpt_inference.transformer.h.16.attn.c_attn.weight", "gpt.gpt_inference.transformer.h.16.attn.c_attn.bias", "gpt.gpt_inference.transformer.h.16.attn.c_proj.weight", "gpt.gpt_inference.transformer.h.16.attn.c_proj.bias", "gpt.gpt_inference.transformer.h.16.ln_2.weight", "gpt.gpt_inference.transformer.h.16.ln_2.bias", "gpt.gpt_inference.transformer.h.16.mlp.c_fc.weight", "gpt.gpt_inference.transformer.h.16.mlp.c_fc.bias", "gpt.gpt_inference.transformer.h.16.mlp.c_proj.weight", "gpt.gpt_inference.transformer.h.16.mlp.c_proj.bias", "gpt.gpt_inference.transformer.h.17.ln_1.weight", "gpt.gpt_inference.transformer.h.17.ln_1.bias", "gpt.gpt_inference.transformer.h.17.attn.c_attn.weight", "gpt.gpt_inference.transformer.h.17.attn.c_attn.bias", "gpt.gpt_inference.transformer.h.17.attn.c_proj.weight", "gpt.gpt_inference.transformer.h.17.attn.c_proj.bias", "gpt.gpt_inference.transformer.h.17.ln_2.weight", "gpt.gpt_inference.transformer.h.17.ln_2.bias", "gpt.gpt_inference.transformer.h.17.mlp.c_fc.weight", "gpt.gpt_inference.transformer.h.17.mlp.c_fc.bias", "gpt.gpt_inference.transformer.h.17.mlp.c_proj.weight", "gpt.gpt_inference.transformer.h.17.mlp.c_proj.bias", "gpt.gpt_inference.transformer.h.18.ln_1.weight", "gpt.gpt_inference.transformer.h.18.ln_1.bias", "gpt.gpt_inference.transformer.h.18.attn.c_attn.weight", "gpt.gpt_inference.transformer.h.18.attn.c_attn.bias", "gpt.gpt_inference.transformer.h.18.attn.c_proj.weight", "gpt.gpt_inference.transformer.h.18.attn.c_proj.bias", "gpt.gpt_inference.transformer.h.18.ln_2.weight", "gpt.gpt_inference.transformer.h.18.ln_2.bias", "gpt.gpt_inference.transformer.h.18.mlp.c_fc.weight", "gpt.gpt_inference.transformer.h.18.mlp.c_fc.bias", "gpt.gpt_inference.transformer.h.18.mlp.c_proj.weight", "gpt.gpt_inference.transformer.h.18.mlp.c_proj.bias", "gpt.gpt_inference.transformer.h.19.ln_1.weight", "gpt.gpt_inference.transformer.h.19.ln_1.bias", "gpt.gpt_inference.transformer.h.19.attn.c_attn.weight", "gpt.gpt_inference.transformer.h.19.attn.c_attn.bias", "gpt.gpt_inference.transformer.h.19.attn.c_proj.weight", "gpt.gpt_inference.transformer.h.19.attn.c_proj.bias", "gpt.gpt_inference.transformer.h.19.ln_2.weight", "gpt.gpt_inference.transformer.h.19.ln_2.bias", "gpt.gpt_inference.transformer.h.19.mlp.c_fc.weight", "gpt.gpt_inference.transformer.h.19.mlp.c_fc.bias", "gpt.gpt_inference.transformer.h.19.mlp.c_proj.weight", "gpt.gpt_inference.transformer.h.19.mlp.c_proj.bias", "gpt.gpt_inference.transformer.h.20.ln_1.weight", "gpt.gpt_inference.transformer.h.20.ln_1.bias", "gpt.gpt_inference.transformer.h.20.attn.c_attn.weight", "gpt.gpt_inference.transformer.h.20.attn.c_attn.bias", "gpt.gpt_inference.transformer.h.20.attn.c_proj.weight", "gpt.gpt_inference.transformer.h.20.attn.c_proj.bias", "gpt.gpt_inference.transformer.h.20.ln_2.weight", "gpt.gpt_inference.transformer.h.20.ln_2.bias", "gpt.gpt_inference.transformer.h.20.mlp.c_fc.weight", "gpt.gpt_inference.transformer.h.20.mlp.c_fc.bias", "gpt.gpt_inference.transformer.h.20.mlp.c_proj.weight", "gpt.gpt_inference.transformer.h.20.mlp.c_proj.bias", "gpt.gpt_inference.transformer.h.21.ln_1.weight", "gpt.gpt_inference.transformer.h.21.ln_1.bias", "gpt.gpt_inference.transformer.h.21.attn.c_attn.weight", "gpt.gpt_inference.transformer.h.21.attn.c_attn.bias", "gpt.gpt_inference.transformer.h.21.attn.c_proj.weight", "gpt.gpt_inference.transformer.h.21.attn.c_proj.bias", "gpt.gpt_inference.transformer.h.21.ln_2.weight", "gpt.gpt_inference.transformer.h.21.ln_2.bias", "gpt.gpt_inference.transformer.h.21.mlp.c_fc.weight", "gpt.gpt_inference.transformer.h.21.mlp.c_fc.bias", "gpt.gpt_inference.transformer.h.21.mlp.c_proj.weight", "gpt.gpt_inference.transformer.h.21.mlp.c_proj.bias", "gpt.gpt_inference.transformer.h.22.ln_1.weight", "gpt.gpt_inference.transformer.h.22.ln_1.bias", "gpt.gpt_inference.transformer.h.22.attn.c_attn.weight", "gpt.gpt_inference.transformer.h.22.attn.c_attn.bias", "gpt.gpt_inference.transformer.h.22.attn.c_proj.weight", "gpt.gpt_inference.transformer.h.22.attn.c_proj.bias", "gpt.gpt_inference.transformer.h.22.ln_2.weight", "gpt.gpt_inference.transformer.h.22.ln_2.bias", "gpt.gpt_inference.transformer.h.22.mlp.c_fc.weight", "gpt.gpt_inference.transformer.h.22.mlp.c_fc.bias", "gpt.gpt_inference.transformer.h.22.mlp.c_proj.weight", "gpt.gpt_inference.transformer.h.22.mlp.c_proj.bias", "gpt.gpt_inference.transformer.h.23.ln_1.weight", "gpt.gpt_inference.transformer.h.23.ln_1.bias", "gpt.gpt_inference.transformer.h.23.attn.c_attn.weight", "gpt.gpt_inference.transformer.h.23.attn.c_attn.bias", "gpt.gpt_inference.transformer.h.23.attn.c_proj.weight", "gpt.gpt_inference.transformer.h.23.attn.c_proj.bias", "gpt.gpt_inference.transformer.h.23.ln_2.weight", "gpt.gpt_inference.transformer.h.23.ln_2.bias", "gpt.gpt_inference.transformer.h.23.mlp.c_fc.weight", "gpt.gpt_inference.transformer.h.23.mlp.c_fc.bias", "gpt.gpt_inference.transformer.h.23.mlp.c_proj.weight", "gpt.gpt_inference.transformer.h.23.mlp.c_proj.bias", "gpt.gpt_inference.transformer.h.24.ln_1.weight", "gpt.gpt_inference.transformer.h.24.ln_1.bias", "gpt.gpt_inference.transformer.h.24.attn.c_attn.weight", "gpt.gpt_inference.transformer.h.24.attn.c_attn.bias", "gpt.gpt_inference.transformer.h.24.attn.c_proj.weight", "gpt.gpt_inference.transformer.h.24.attn.c_proj.bias", "gpt.gpt_inference.transformer.h.24.ln_2.weight", "gpt.gpt_inference.transformer.h.24.ln_2.bias", "gpt.gpt_inference.transformer.h.24.mlp.c_fc.weight", "gpt.gpt_inference.transformer.h.24.mlp.c_fc.bias", "gpt.gpt_inference.transformer.h.24.mlp.c_proj.weight", "gpt.gpt_inference.transformer.h.24.mlp.c_proj.bias", "gpt.gpt_inference.transformer.h.25.ln_1.weight", "gpt.gpt_inference.transformer.h.25.ln_1.bias", "gpt.gpt_inference.transformer.h.25.attn.c_attn.weight", "gpt.gpt_inference.transformer.h.25.attn.c_attn.bias", "gpt.gpt_inference.transformer.h.25.attn.c_proj.weight", "gpt.gpt_inference.transformer.h.25.attn.c_proj.bias", "gpt.gpt_inference.transformer.h.25.ln_2.weight", "gpt.gpt_inference.transformer.h.25.ln_2.bias", "gpt.gpt_inference.transformer.h.25.mlp.c_fc.weight", "gpt.gpt_inference.transformer.h.25.mlp.c_fc.bias", "gpt.gpt_inference.transformer.h.25.mlp.c_proj.weight", "gpt.gpt_inference.transformer.h.25.mlp.c_proj.bias", "gpt.gpt_inference.transformer.h.26.ln_1.weight", "gpt.gpt_inference.transformer.h.26.ln_1.bias", "gpt.gpt_inference.transformer.h.26.attn.c_attn.weight", "gpt.gpt_inference.transformer.h.26.attn.c_attn.bias", "gpt.gpt_inference.transformer.h.26.attn.c_proj.weight", "gpt.gpt_inference.transformer.h.26.attn.c_proj.bias", "gpt.gpt_inference.transformer.h.26.ln_2.weight", "gpt.gpt_inference.transformer.h.26.ln_2.bias", "gpt.gpt_inference.transformer.h.26.mlp.c_fc.weight", "gpt.gpt_inference.transformer.h.26.mlp.c_fc.bias", "gpt.gpt_inference.transformer.h.26.mlp.c_proj.weight", "gpt.gpt_inference.transformer.h.26.mlp.c_proj.bias", "gpt.gpt_inference.transformer.h.27.ln_1.weight", "gpt.gpt_inference.transformer.h.27.ln_1.bias", "gpt.gpt_inference.transformer.h.27.attn.c_attn.weight", "gpt.gpt_inference.transformer.h.27.attn.c_attn.bias", "gpt.gpt_inference.transformer.h.27.attn.c_proj.weight", "gpt.gpt_inference.transformer.h.27.attn.c_proj.bias", "gpt.gpt_inference.transformer.h.27.ln_2.weight", "gpt.gpt_inference.transformer.h.27.ln_2.bias", "gpt.gpt_inference.transformer.h.27.mlp.c_fc.weight", "gpt.gpt_inference.transformer.h.27.mlp.c_fc.bias", "gpt.gpt_inference.transformer.h.27.mlp.c_proj.weight", "gpt.gpt_inference.transformer.h.27.mlp.c_proj.bias", "gpt.gpt_inference.transformer.h.28.ln_1.weight", "gpt.gpt_inference.transformer.h.28.ln_1.bias", "gpt.gpt_inference.transformer.h.28.attn.c_attn.weight", "gpt.gpt_inference.transformer.h.28.attn.c_attn.bias", "gpt.gpt_inference.transformer.h.28.attn.c_proj.weight", "gpt.gpt_inference.transformer.h.28.attn.c_proj.bias", "gpt.gpt_inference.transformer.h.28.ln_2.weight", "gpt.gpt_inference.transformer.h.28.ln_2.bias", "gpt.gpt_inference.transformer.h.28.mlp.c_fc.weight", "gpt.gpt_inference.transformer.h.28.mlp.c_fc.bias", "gpt.gpt_inference.transformer.h.28.mlp.c_proj.weight", "gpt.gpt_inference.transformer.h.28.mlp.c_proj.bias", "gpt.gpt_inference.transformer.h.29.ln_1.weight", "gpt.gpt_inference.transformer.h.29.ln_1.bias", "gpt.gpt_inference.transformer.h.29.attn.c_attn.weight", "gpt.gpt_inference.transformer.h.29.attn.c_attn.bias", "gpt.gpt_inference.transformer.h.29.attn.c_proj.weight", "gpt.gpt_inference.transformer.h.29.attn.c_proj.bias", "gpt.gpt_inference.transformer.h.29.ln_2.weight", "gpt.gpt_inference.transformer.h.29.ln_2.bias", "gpt.gpt_inference.transformer.h.29.mlp.c_fc.weight", "gpt.gpt_inference.transformer.h.29.mlp.c_fc.bias", "gpt.gpt_inference.transformer.h.29.mlp.c_proj.weight", "gpt.gpt_inference.transformer.h.29.mlp.c_proj.bias", "gpt.gpt_inference.transformer.ln_f.weight", "gpt.gpt_inference.transformer.ln_f.bias", "gpt.gpt_inference.transformer.wte.weight", "gpt.gpt_inference.pos_embedding.emb.weight", "gpt.gpt_inference.embeddings.weight", "gpt.gpt_inference.final_norm.weight", "gpt.gpt_inference.final_norm.bias", "gpt.gpt_inference.lm_head.0.weight", "gpt.gpt_inference.lm_head.0.bias", "gpt.gpt_inference.lm_head.1.weight", "gpt.gpt_inference.lm_head.1.bias".
Unexpected key(s) in state_dict: "gpt.conditioning_perceiver.latents", "gpt.conditioning_perceiver.layers.0.0.to_q.weight", "gpt.conditioning_perceiver.layers.0.0.to_kv.weight", "gpt.conditioning_perceiver.layers.0.0.to_out.weight", "gpt.conditioning_perceiver.layers.0.1.0.weight", "gpt.conditioning_perceiver.layers.0.1.0.bias", "gpt.conditioning_perceiver.layers.0.1.2.weight", "gpt.conditioning_perceiver.layers.0.1.2.bias", "gpt.conditioning_perceiver.layers.1.0.to_q.weight", "gpt.conditioning_perceiver.layers.1.0.to_kv.weight", "gpt.conditioning_perceiver.layers.1.0.to_out.weight", "gpt.conditioning_perceiver.layers.1.1.0.weight", "gpt.conditioning_perceiver.layers.1.1.0.bias", "gpt.conditioning_perceiver.layers.1.1.2.weight", "gpt.conditioning_perceiver.layers.1.1.2.bias", "gpt.conditioning_perceiver.norm.gamma".
size mismatch for gpt.mel_embedding.weight: copying a param with shape torch.Size([1026, 1024]) from checkpoint, the shape in current model is torch.Size([8194, 1024]).
size mismatch for gpt.mel_head.weight: copying a param with shape torch.Size([1026, 1024]) from checkpoint, the shape in current model is torch.Size([8194, 1024]).
size mismatch for gpt.mel_head.bias: copying a param with shape torch.Size([1026]) from checkpoint, the shape in current model is torch.Size([8194]).
```
### Environment
```shell
{
"CUDA": {
"GPU": ["NVIDIA A100-SXM4-80GB"],
"available": true,
"version": "11.8"
},
"Packages": {
"PyTorch_debug": false,
"PyTorch_version": "2.1.0+cu118",
"TTS": "0.20.1",
"numpy": "1.22.0"
},
"System": {
"OS": "Linux",
"architecture": [
"64bit",
"ELF"
],
"processor": "x86_64",
"python": "3.9.18",
"version": "#183-Ubuntu SMP Mon Oct 2 11:28:33 UTC 2023"
}
}
```
### Additional context
_No response_
|
closed
|
2023-11-09T03:28:30Z
|
2024-06-25T12:46:25Z
|
https://github.com/coqui-ai/TTS/issues/3177
|
[
"bug"
] |
caffeinetoomuch
| 12
|
ultralytics/yolov5
|
pytorch
| 13,243
|
Exporting trained yolov5 model (trained on custom dataset) to 'saved model' format changes the no. of classes and the name of classes to default coco128 values
|
### Search before asking
- [X] I have searched the YOLOv5 [issues](https://github.com/ultralytics/yolov5/issues) and found no similar bug report.
### YOLOv5 Component
Export
### Bug
I trained yolov5s model to detect various logos (amazon, ups, fedex etc). The model detects the logos well.
The command used for training is:
```python train.py --weights yolov5s.pt --epoch 100 --data C:\projects\logo_detector\yolov5\datasetv3\data.yaml```
The command used for detecting logos is:
```python detect.py --weights best.pt --source 0```
Screenshot of trained yolov5s model detecting the logos:

When I use export.py to convert the above model to saved model format, the model starts giving wrong output.
The command used for exporting the model is:
```python export.py --weights best.pt --data C:\projects\logo_detector\yolov5\datasetv3\data.yaml --include saved_model```
The command used for detection of logos using this saved model is:
```python detect.py --weights best_saved_model --source 0```
Screenshot of yolov5s saved model giving wrong output is:

As far as I can understand, the model starts giving output according to the default coco128.yaml file. But I have not specified this file in my commands, so I cannot understand the reason behind this behaviour. Please let me know how to get correct output.
### Environment
- I have used the default git repository for yolov5
- OS: Windows 10 Pro
- Python: 3.12.3
### Minimal Reproducible Example
_No response_
### Additional
_No response_
### Are you willing to submit a PR?
- [ ] Yes I'd like to help by submitting a PR!
|
open
|
2024-08-05T07:38:31Z
|
2024-10-27T13:30:48Z
|
https://github.com/ultralytics/yolov5/issues/13243
|
[
"bug"
] |
ssingh17j
| 2
|
Lightning-AI/pytorch-lightning
|
deep-learning
| 19,978
|
Running `test` with LightningCLI, the program can quit before the test loop ends
|
### Bug description
Within my `LightningModule`, I used `self.log_dict(metrics, on_step=True, on_epoch=True)` in `test_step`, and run with `python main.py test --config config.yaml`, with `main.py` containing only `cli = LightningCLI()`, and `config.yaml` providing both the datasets and model. The `TensorBoardLogger` is used.
However, after the programs ends, sometimes I can normally get the metrics `epoch`, `test_accuracy_epoch` and `test_loss_epoch` in the logger file, but at most attempts these 3 metrics didn't show up, and step-level logged objects can always be seen normally.
When the problems occurs, nothing abnormal can be seen from command line outputs. It looks as if the program quited normally.
I find a walkaround to be sleeping for a while in `main.py` right after `cli = LightningCLI()`. It seems like this is because a child thread is not waited to the end.
### What version are you seeing the problem on?
v2.2
### How to reproduce the bug
main.py
```python
from lightning.pytorch.cli import LightningCLI
from lightning.pytorch.loggers import TensorBoardLogger
from lightning.pytorch.callbacks import ModelCheckpoint
from model import Model
from datamodule import DataModule
def cli_main():
cli = LightningCLI()
if __name__ == "__main__":
cli_main()
from time import sleep
sleep(2)
# The problem can be solved by adding sleep.
```
config.yaml
```yaml
# lightning.pytorch==2.2.5
ckpt_path: null
seed_everything: 0
model:
class_path: model.Model
init_args:
learning_rate: 1e-3
data:
class_path: datamodule.DataModule
init_args:
data_dir: data
trainer:
accelerator: gpu
strategy: auto
devices: 1
num_nodes: 1
precision: null
fast_dev_run: false
max_epochs: 100
min_epochs: null
max_steps: -1
min_steps: null
max_time: null
limit_train_batches: null
limit_val_batches: 10
limit_test_batches: null
limit_predict_batches: null
logger:
class_path: lightning.pytorch.loggers.TensorBoardLogger
init_args:
save_dir: lightning_logs/resnet50
name: normalized
callbacks:
class_path: lightning.pytorch.callbacks.ModelCheckpoint
init_args:
save_top_k: 5
monitor: valid_loss
filename: "{epoch}-{step}-{valid_loss:.8f}"
overfit_batches: 0.0
val_check_interval: 50
check_val_every_n_epoch: 1
num_sanity_val_steps: null
log_every_n_steps: 50
enable_checkpointing: null
enable_progress_bar: null
enable_model_summary: null
accumulate_grad_batches: 1
gradient_clip_val: null
gradient_clip_algorithm: null
deterministic: false
benchmark: null
inference_mode: true
use_distributed_sampler: true
profiler: null
detect_anomaly: false
barebones: false
plugins: null
sync_batchnorm: true
reload_dataloaders_every_n_epochs: 0
default_root_dir: null
```
model.py
```Python
import torch
from torch import nn
import torch.nn.functional as F
import lightning as pl
from torchvision.models import resnet50
class Model(pl.LightningModule):
def __init__(self, learning_rate: float):
super().__init__()
self.save_hyperparameters()
CHARS = "0123456789ABCDEFGHIJKLMNOPQRSTUVWXYZabcdefghijklmnopqrstuvwxyz"
class_num = len(CHARS)
self.text_len = 4
resnet = resnet50()
resnet.conv1 = nn.Conv2d(
1, 64, kernel_size=(7, 7), stride=(2, 2), padding=(3, 3), bias=False
)
layers = list(resnet.children())
self.resnet = nn.Sequential(*layers[:9])
self.linear = nn.Linear(512, class_num)
self.softmax = nn.Softmax(2)
def _calc_softmax(self, x: torch.Tensor) -> torch.Tensor:
x = self.resnet(x) # (batch, 2048, 1, 1)
x = x.reshape(x.shape[0], self.text_len, -1) # (batch, 4, 512)
x = self.linear(x) # (batch, 4, 62)
x = self.softmax(x) # (batch, 4, 62)
return x
def forward(self, x: torch.Tensor) -> torch.Tensor:
# in lightning, forward defines the prediction/inference actions
x = self._calc_softmax(x) # (batch, 4, 62)
return torch.argmax(x, 2) # (batch, 4)
def training_step(self, batch: torch.Tensor, batch_idx: int) -> torch.Tensor:
# training_step defined the train loop.
# It is independent of forward
img, target = batch
batch_size = img.shape[0]
pred_softmax = self._calc_softmax(img) # (batch, 4, 62)
pred_softmax_permute = pred_softmax.permute((0, 2, 1)) # (batch, 62, 4)
loss = F.cross_entropy(pred_softmax_permute, target)
with torch.no_grad():
pred = torch.argmax(pred_softmax, 2) # (batch, 4)
char_correct = (pred == target).sum(1) # (batch)
batch_correct = (char_correct == self.text_len).sum()
batch_accuracy = batch_correct / batch_size
metrics = {"train_accuracy": batch_accuracy, "train_loss": loss}
self.log_dict(metrics, prog_bar=True, logger=True, on_step=True, on_epoch=True)
return loss
def configure_optimizers(self) -> torch.optim.Optimizer:
optimizer = torch.optim.Adam(self.parameters(), lr=self.hparams.learning_rate)
return optimizer
def validation_step(self, batch: torch.Tensor, batch_idx: int) -> torch.Tensor:
# validation_step defined the validation loop.
# It is independent of forward
img, target = batch
batch_size = img.shape[0]
pred_softmax = self._calc_softmax(img) # (batch, 4, 62)
pred_softmax_permute = pred_softmax.permute((0, 2, 1)) # (batch, 62, 4)
loss = F.cross_entropy(pred_softmax_permute, target)
with torch.no_grad():
pred = torch.argmax(pred_softmax, 2) # (batch, 4)
char_correct = (pred == target).sum(1) # (batch)
batch_correct = (char_correct == self.text_len).sum()
batch_accuracy = batch_correct / batch_size
metrics = {"valid_accurary": batch_accuracy, "valid_loss": loss}
self.log_dict(metrics, prog_bar=True, logger=True, on_step=True, on_epoch=True)
return loss
def test_step(self, batch: torch.Tensor, batch_idx: int) -> torch.Tensor:
# test_step defined the test loop.
# It is independent of forward
img, target = batch
batch_size = img.shape[0]
pred_softmax = self._calc_softmax(img) # (batch, 4, 62)
pred_softmax_permute = pred_softmax.permute((0, 2, 1)) # (batch, 62, 4)
loss = F.cross_entropy(pred_softmax_permute, target)
with torch.no_grad():
pred = torch.argmax(pred_softmax, 2) # (batch, 4)
char_correct = (pred == target).sum(1) # (batch)
batch_correct = (char_correct == self.text_len).sum()
batch_accuracy = batch_correct / batch_size
metrics = {"test_accurary": batch_accuracy, "test_loss": loss}
self.log_dict(metrics, prog_bar=True, logger=True, on_step=True, on_epoch=True)
## The `on_epoch` part of behaviors are unstable, but `test_accuracy_step` can always be seen.
## If `on_step=False` and `on_epoch=True`, it works fine to me.
return loss
```
```
### Error messages and logs
```
# Error messages and logs here please
```
### Environment
<details>
<summary>Current environment</summary>
```
- PyTorch Lightning Version: 2.2.5
- PyTorch Version: 2.3.1+cu121
- Python version: 3.12.4
- OS: Windows 11
- CUDA/cuDNN version: 12.1
- GPU models and configuration: GTX 1650
- How you installed Lightning: pip
```
</details>
### More info
_No response_
|
open
|
2024-06-15T16:24:26Z
|
2024-06-15T16:36:11Z
|
https://github.com/Lightning-AI/pytorch-lightning/issues/19978
|
[
"bug",
"needs triage"
] |
t4rf9
| 0
|
autogluon/autogluon
|
computer-vision
| 4,792
|
[timeseries] Clarify the documentation for `known_covariates` during `predict()`
|
## Description
We should clarify which values should be provided as `known_covariates` during prediction time.
The [current documentation](https://auto.gluon.ai/stable/tutorials/timeseries/forecasting-indepth.html) says:
"The timestamp index must include the values for prediction_length many time steps into the future from the end of each time series in train_data".
This formulation is ambiguous.
- [ ] impove wording in the documentation
- [ ] add a method `TimeSeriesPredictor.get_forecast_index(data) -> pd.MultiIndex` that returns the `item_id, timestamp` index that should be covered by the `known_covariates` during `predict()`.
|
open
|
2025-01-14T08:03:33Z
|
2025-01-14T08:05:06Z
|
https://github.com/autogluon/autogluon/issues/4792
|
[
"API & Doc",
"enhancement",
"module: timeseries"
] |
shchur
| 0
|
pyeve/eve
|
flask
| 711
|
extra_response_fields should be after (not before) any on_inserted hooks on POST
|
Currently, `extra_response_fields` are processed after `on_insert` hooks are complete but before any `on_inserted` hooks.
It would be intuitive and great to have `extra_response_fields` processed after both of these hooks are complete - in case we changed something during `on_inserted`.
|
closed
|
2015-09-14T05:20:29Z
|
2018-05-18T18:19:30Z
|
https://github.com/pyeve/eve/issues/711
|
[
"enhancement",
"on hold",
"stale"
] |
kenmaca
| 2
|
DistrictDataLabs/yellowbrick
|
scikit-learn
| 949
|
Some plot directive visualizers not rendering in Read the Docs
|
Currently on Read the Docs (develop branch), a few of our visualizers that use the plot directive (#687) are not rendering the plots:
- [Classification Report](http://www.scikit-yb.org/en/develop/api/classifier/classification_report.html)
- [Silhouette Scores](http://www.scikit-yb.org/en/develop/api/cluster/silhouette.html)
- [ScatterPlot](http://www.scikit-yb.org/en/develop/api/contrib/scatter.html)
- [JointPlot](http://www.scikit-yb.org/en/develop/api/features/jointplot.html)
|
closed
|
2019-08-15T20:58:39Z
|
2019-08-29T00:03:24Z
|
https://github.com/DistrictDataLabs/yellowbrick/issues/949
|
[
"type: bug",
"type: documentation"
] |
rebeccabilbro
| 1
|
plotly/dash
|
flask
| 2,754
|
[BUG] Dropdown options not rendering on the UI even though it is generated
|
**Describe your context**
Python Version -> `3.8.18`
`poetry show | grep dash` gives the below packages:
```
dash 2.7.0 A Python framework for building reac...
dash-bootstrap-components 1.5.0 Bootstrap themed components for use ...
dash-core-components 2.0.0 Core component suite for Dash
dash-html-components 2.0.0 Vanilla HTML components for Dash
dash-prefix 0.0.4 Dash library for managing component IDs
dash-table 5.0.0 Dash table
```
- if frontend related, tell us your Browser, Version and OS
- OS: MacOSx (Sonoma 14.3)
- Browser: Chrome (also tried on Firefox and Safari)
- Version: 121.0.6167.160 (Official Build) (x86_64)
**Describe the bug**
I have a multi-dropdown that syncs up with the input from a separate tab to pull in the list of regions associated with a country. A particular country, GB, when selected does not seem to populate the dropdown options. The UI element created was written to stdout which lists the elements correctly, but it does not render on the UI itself.
stdout printout is as follows:
```
Div([P(children='Group A - (Control)', style={'marginBottom': 5}),
Dropdown(options=[
{'label': 'Cheshire', 'value': 'Cheshire'},
{'label': 'Leicestershire', 'value': 'Leicestershire'},
{'label': 'Hertfordshire', 'value': 'Hertfordshire'},
{'label': 'Surrey', 'value': 'Surrey'},
{'label': 'Lancashire', 'value': 'Lancashire'},
{'label': 'Warwickshire', 'value': 'Warwickshire'},
{'label': 'Cumbria', 'value': 'Cumbria'},
{'label': 'Northamptonshire', 'value': 'Northamptonshire'},
{'label': 'Dorset', 'value': 'Dorset'},
{'label': 'Isle of Wight', 'value': 'Isle of Wight'},
{'label': 'Kent', 'value': 'Kent'},
{'label': 'Lincolnshire', 'value': 'Lincolnshire'},
{'label': 'Hampshire', 'value': 'Hampshire'},
{'label': 'Cornwall', 'value': 'Cornwall'},
{'label': 'Scotland', 'value': 'Scotland'},
{'label': 'Berkshire', 'value': 'Berkshire'},
{'label': 'Gloucestershire, Wiltshire & Bristol', 'value': 'Gloucestershire, Wiltshire & Bristol'},
{'label': 'Durham', 'value': 'Durham'},
{'label': 'Rutland', 'value': 'Rutland'},
{'label': 'Northumberland', 'value': 'Northumberland'},
{'label': 'West Midlands', 'value': 'West Midlands'},
{'label': 'Derbyshire', 'value': 'Derbyshire'},
{'label': 'Merseyside', 'value': 'Merseyside'},
{'label': 'East Sussex', 'value': 'East Sussex'},
{'label': 'Northern Ireland', 'value': 'Northern Ireland'},
{'label': 'Oxfordshire', 'value': 'Oxfordshire'},
{'label': 'Herefordshire', 'value': 'Herefordshire'},
{'label': 'Staffordshire', 'value': 'Staffordshire'},
{'label': 'East Riding of Yorkshire', 'value': 'East Riding of Yorkshire'},
{'label': 'South Yorkshire', 'value': 'South Yorkshire'},
{'label': 'West Sussex', 'value': 'West Sussex'},
{'label': 'Tyne and Wear', 'value': 'Tyne and Wear'},
{'label': 'Buckinghamshire', 'value': 'Buckinghamshire'},
{'label': 'West Yorkshire', 'value': 'West Yorkshire'},
{'label': 'Wales', 'value': 'Wales'},
{'label': 'Somerset', 'value': 'Somerset'},
{'label': 'Worcestershire', 'value': 'Worcestershire'},
{'label': 'North Yorkshire', 'value': 'North Yorkshire'},
{'label': 'Shropshire', 'value': 'Shropshire'},
{'label': 'Nottinghamshire', 'value': 'Nottinghamshire'},
{'label': 'Essex', 'value': 'Essex'},
{'label': 'Greater London & City of London', 'value': 'Greater London & City of London'},
{'label': 'Cambridgeshire', 'value': 'Cambridgeshire'},
{'label': 'Greater Manchester', 'value': 'Greater Manchester'},
{'label': 'Suffolk', 'value': 'Suffolk'},
{'label': 'Norfolk', 'value': 'Norfolk'},
{'label': 'Devon', 'value': 'Devon'},
{'label': 'Bedfordshire', 'value': 'Bedfordshire'}],
value=[],
multi=True,
id={'role': 'experiment-design-geoassignment-manual-geodropdown', 'group_id': 'Group-ID1234'})])
```
**Expected behavior**
When the country GB is selected, I expect the relevant options to be populated in the dropdown that can be selected. The code below:
``` python
def get_geos(self, all_geos):
element = html.Div(
[
html.P("TEST", style={"marginBottom": 5}),
dcc.Dropdown(
id={"role": self.prefix("dropdown"), "group_id": "1234"},
multi=True,
value=[],
searchable=True,
options=[{"label": g, "value": g} for g in all_geos],
),
]
)
print(element) # Print output is posted above showing that the callback is working fine. But it is not rendering correctly on the front end
return element
```
**Screen Recording**
https://github.com/plotly/dash/assets/94958897/13909683-244c-4cbe-853a-be148f3aae1c
|
closed
|
2024-02-08T13:47:01Z
|
2024-05-31T20:12:51Z
|
https://github.com/plotly/dash/issues/2754
|
[] |
malavika-menon
| 2
|
nikitastupin/clairvoyance
|
graphql
| 100
|
500 internal server error
|
Hey tool showing 500 ERROR on loop, i then burp Intercepted my clairvoyance traffic
clairvoyance -H "Authorization: Bearer" -H "X-api-key:" -x "127.1:8080" -k http://example.com/graphql
**Body it sending**
`{"query": "query { reporting essential myself tours platform load affiliate labor immediately admin nursing defense machines designated tags heavy covered recovery joe guys integrated configuration merchant comprehensive expert universal protect drop solid cds presentation languages became orange compliance vehicles prevent theme rich im campaign marine improvement vs guitar finding pennsylvania examples ipod saying spirit ar claims challenge motorola acceptance strategies mo seem affairs touch intended towards sa }"}`
**Response**
```
HTTP/2 500 Internal Server Error
Content-Type: application/json; charset=utf-8
{"errors":[{"message":"Too many validation errors, error limit reached. Validation aborted.","extensions":{"code":"INTERNAL_SERVER_ERROR"}}]}
```
but sending manually this it works:
`{"query": "query { along among death writing speed }"}`
|
open
|
2024-05-15T15:16:51Z
|
2024-08-27T06:13:53Z
|
https://github.com/nikitastupin/clairvoyance/issues/100
|
[
"bug"
] |
649abhinav
| 1
|
predict-idlab/plotly-resampler
|
plotly
| 341
|
Dash Callback says FigureResampler is not JSON serializable
|
Apologies, this is more of a "this broke and I don't know what went wrong" type of issue. What it looks like so far is that everything in the dash dashboard ive made works except for the plotting. This is the exception i get:
```
dash.exceptions.InvalidCallbackReturnValue: The callback for `[<Output `data-plot.figure`>, <Output `store.data`>, <Output `status-msg.children`>]`
returned a value having type `FigureResampler`
which is not JSON serializable.
The value in question is either the only value returned,
or is in the top level of the returned list,
and has string representation
`FigureResampler({ 'data': [{'mode': 'lines',
'name': '<b style="color:sandybrown">[R]</b> Category1 <i style="color:#fc9944">~10s</i>',
'type': 'scatter',
'uid': 'c78a3bb2-658c-44d0-b791-dfc0bbe76cd8',
'x': array([datetime.datetime(2025, 1, 22, 18, 38, 21),...
```
The relevant code chunks that could cause this break is:
```
from dash import Dash, html, dcc, Output, Input, State, callback, no_update, ctx
from dash_extensions.enrich import DashProxy, ServersideOutputTransform, Serverside
import dash_bootstrap_components as dbc
import pandas as pd
import plotly.express as px
app = DashProxy(
__name__,
external_stylesheets=[dbc.themes.LUX],
transforms=[ServersideOutputTransform()],
)
# assume app creation within a dbc. container here
dcc.Graph(id="data-plot", figure=go.Figure())
# this is the callback for the function triggering the break:
@callback(
[Output("data-plot", "figure"),
Output("store", "data"), # Cache the figure data
Output("status-msg", "children")],
[Input("load-btn", "n_clicks"),
State("dropdown-1", "value"),
State("dropdown-2", "value"),
State("dropdown-3", "value"),
State("dropdown-4", "value"),
State("dropdown-5", "value")],
prevent_initial_call=True # Prevents callback from running at startup
)
# this is how i made the figure, assume its right next to the call back from above
fig = FigureResampler(go.Figure(), default_n_shown_samples=10000)
# added trace and update layout here
# and then i return fig, Serverside(fig), "this thing works"
# i also use this function to update the resampling
@app.callback(
Output("data-plot", "figure", allow_duplicate=True),
Input("data-plot", "relayoutData"),
State("store", "data"), # The server side cached FigureResampler per session
prevent_initial_call=True,
)
def update_fig(relayoutdata: dict, fig: FigureResampler):
if fig is None:
return no_update
return fig.construct_update_data_patch(relayoutdata)
```
it looks like from the docs that you can return plotly resample figures as returns to the output for dcc.Graph. what could have gone wrong?
|
closed
|
2025-03-05T20:37:21Z
|
2025-03-06T18:06:15Z
|
https://github.com/predict-idlab/plotly-resampler/issues/341
|
[] |
FDSRashid
| 1
|
matplotlib/matplotlib
|
matplotlib
| 29,799
|
[ENH]: set default color cycle to named color sequence
|
### Problem
It would be great if I could put something like this in my matplotlibrc to use the petroff10 color sequence by default:
```
axes.prop_cycle : cycler('color', 'petroff10')
```
### Proposed solution
Currently if a single string is supplied we try to interpret as a list of single character colors
https://github.com/matplotlib/matplotlib/blob/a9dc9acc2dd1bab761b45e48c8d63aa108811a82/lib/matplotlib/rcsetup.py#L105-L108
None of the current built in color sequences can be interpreted that way, so it would not be ambiguous to try both that and querying the color sequence registry. However, maybe we would need something to guard against user- or third-party-defined color sequences having a name like "rygbk"?
|
open
|
2025-03-24T16:57:39Z
|
2025-03-24T17:42:14Z
|
https://github.com/matplotlib/matplotlib/issues/29799
|
[
"New feature",
"topic: rcparams",
"topic: color/cycle"
] |
rcomer
| 2
|
horovod/horovod
|
pytorch
| 3,795
|
Seen with tf-head: ModuleNotFoundError: No module named 'keras.optimizers.optimizer_v2'
|
Problem with tf-head / Keras seen in CI, for instance at https://github.com/horovod/horovod/actions/runs/3656223581/jobs/6180240570
```
___________ ERROR collecting test/parallel/test_tensorflow_keras.py ____________
ImportError while importing test module '/horovod/test/parallel/test_tensorflow_keras.py'.
Hint: make sure your test modules/packages have valid Python names.
Traceback:
/usr/lib/python3.8/importlib/__init__.py:127: in import_module
return _bootstrap._gcd_import(name[level:], package, level)
test_tensorflow_keras.py:36: in <module>
from keras.optimizers.optimizer_v2 import optimizer_v2
E ModuleNotFoundError: No module named 'keras.optimizers.optimizer_v2'
```
```
___________ ERROR collecting test/parallel/test_tensorflow2_keras.py ___________
ImportError while importing test module '/horovod/test/parallel/test_tensorflow2_keras.py'.
Hint: make sure your test modules/packages have valid Python names.
Traceback:
/usr/lib/python3.8/importlib/__init__.py:127: in import_module
return _bootstrap._gcd_import(name[level:], package, level)
test_tensorflow2_keras.py:35: in <module>
from keras.optimizers.optimizer_v2 import optimizer_v2
E ModuleNotFoundError: No module named 'keras.optimizers.optimizer_v2'
```
|
closed
|
2022-12-09T14:25:50Z
|
2022-12-10T09:54:00Z
|
https://github.com/horovod/horovod/issues/3795
|
[
"bug"
] |
maxhgerlach
| 1
|
plotly/dash-table
|
plotly
| 700
|
`Backspace` on cell only reflects deleted content after cell selection changes
|
In the recording below, `backspace` is hit right after the cell selection and the displayed cell content only updates after the selected cell changed.

|
open
|
2020-02-20T17:08:28Z
|
2024-01-25T21:34:23Z
|
https://github.com/plotly/dash-table/issues/700
|
[
"dash-type-bug",
"regression"
] |
Marc-Andre-Rivet
| 2
|
AirtestProject/Airtest
|
automation
| 1,205
|
airtest自动安装的urllib3库, 需要旧版 (比如1.26.17) 才能通过uid连接ios手机
|
**描述问题bug**
airtest自动安装的urllib3库, 需要旧版 (比如1.26.17) 才能通过uuid连接ios手机, 否则会提示wda未准备好并且在20秒等待后报错, 当你将 urllib3库改为旧版本可以解决这个问题, 控制端mac和windows 设备端ios15/16/17 下均是如此.
**python 版本:** `python3.11`
**airtest 版本:** `1.3.3`
**设备:**
- 手机型号: [iphone se2]
- 控制端: [mbp m1/windows11]
- 手机系统: [ios15/ios16/ios17]
|
open
|
2024-04-15T06:33:57Z
|
2024-04-15T06:41:39Z
|
https://github.com/AirtestProject/Airtest/issues/1205
|
[] |
yh1121yh
| 0
|
ultrafunkamsterdam/undetected-chromedriver
|
automation
| 2,155
|
[NODRIVER] Add ability to capture and return screenshot as base64 - changes are ready to PR merge
|
Hello, I would like to create a PR to (as title suggest) giving the base64 of the screenshots instead of saving files locally
Here is the commit: https://github.com/falmar/nodriver/commit/d903cca8aac2406ff0c4462785b61d5ce474256c
it includes and demo example
EDIT: I'm unable to create PR on the nodriver [repository](https://github.com/ultrafunkamsterdam/nodriver)
|
closed
|
2025-03-07T12:29:58Z
|
2025-03-10T07:48:53Z
|
https://github.com/ultrafunkamsterdam/undetected-chromedriver/issues/2155
|
[] |
falmar
| 2
|
xinntao/Real-ESRGAN
|
pytorch
| 209
|
希望能增加带去除扫描图片网纹的超分辨率算法
|
在进行扫图的杂志 周边 同人志等超分辨率的时候 网纹也会被放大得非常明显 不知道有没有办法先把网纹去除后再进行超分辨率呢?
|
open
|
2022-01-02T15:53:01Z
|
2022-01-02T15:53:01Z
|
https://github.com/xinntao/Real-ESRGAN/issues/209
|
[] |
sistinafibe
| 0
|
2noise/ChatTTS
|
python
| 3
|
运行到一半就自动停止了
|

如图,运行到一半就停止了。。
系统:linux
python版本:3.12
另外建议写个requirements吧
|
closed
|
2024-05-28T02:50:19Z
|
2024-05-28T11:29:23Z
|
https://github.com/2noise/ChatTTS/issues/3
|
[] |
luosaidage
| 1
|
kizniche/Mycodo
|
automation
| 442
|
install mycodo error???
|
what happen ?! How to install mycodo version lower to 5.6.10? I'm want to install mycodo version 5.5.24. how can i do?
|
closed
|
2018-04-03T07:11:43Z
|
2018-04-06T00:42:52Z
|
https://github.com/kizniche/Mycodo/issues/442
|
[] |
bike2538
| 4
|
InstaPy/InstaPy
|
automation
| 6,296
|
Not posting the comment when mentioning any account.
|
## InstaPy configuration
InstaPy Version: 0.6.14-AS
I am trying to comment on a hashtag by mentioning some page with @..... but it seems like when I am using the @.... it doesn't post the comment instead it skips it. Anyone else is facing the same issue? What is the solution?
|
open
|
2021-08-16T09:16:11Z
|
2021-10-10T00:35:52Z
|
https://github.com/InstaPy/InstaPy/issues/6296
|
[] |
moshema10
| 6
|
StackStorm/st2
|
automation
| 6,160
|
Provide support for passing "=" in a string
|
**alias yaml**
`---
name: "launch_quasar"
action_ref: "quasar.quasar1"
description: "launch a quasar execution"
formats:
- display: "*<command>* *<payload>*"
representation:
- "{{ command }} {{ payload }}"
result:
format: |
```{{ execution.result.result }}```
`
**action yaml**
name: quasar1
description: Action that takes an input parameter
runner_type: 'python-script'
entry_point: 'quasar1.py'
enabled: true
parameters:
command:
type: string
description: 'Input parameter'
required: true
payload:
type: string
description: 'Input parameter'
required: true
user:
type: "string"
description: "Slack user who triggered the action"
required: false
default: "{{action_context.api_user}}"
**Representation used:** "{{ command }} {{ payload }}"
**Parameter defintion**:
payload:
type: string
description: 'Input parameter'
required: true
We have an usecase where we need to pass a string with "=" in it for payload for some reason stackstorm is not allowing me to do that, if i pass such value it is not taking it as a seperate string and causing multiple issues
Issue is not oberved if we are putting ":" instead of "=" and also putting a space after "=" solves the issue
Have tried different things (using jinja template replace option and replaced "=" with "= " but i am getting internal server error.
if i am able to pass the yaml validation, i can have my own validation in action py file but the issue the execution is not even going to the python file.
basically this should be accepted by stackstorm **quasar create cluster_name=weekly_nats**
<img width="372" alt="Screenshot 2024-03-04 at 11 35 28 AM" src="https://github.com/StackStorm/st2/assets/41072130/ae316000-c84a-4493-976e-e2e6f5b38fbc">
|
open
|
2024-03-04T06:08:03Z
|
2024-03-04T06:09:32Z
|
https://github.com/StackStorm/st2/issues/6160
|
[] |
sivudu47
| 0
|
assafelovic/gpt-researcher
|
automation
| 645
|
UnicodeEncodeError: 'charmap' codec can't encode character '\U0001f50e' in position 0: character maps to <undefined>
|
I'm testing a simple next.js/fastapi app on Windows 11, using the example FastAPI from https://docs.tavily.com/docs/gpt-researcher/pip-package (btw I think this example is missing the query parameter)
It's a very simple parameter report test with api call/url of
```
const query = encodeURIComponent("What is 42?");
const type = encodeURIComponent("outline_report");
const URL = `/api/reporttest/${query}/${type}`;
const URL2 = `/api/report/${query}/${type}`;
```
URL is a simple parameter test
```
@app.get("/api/reporttest/{query}/{report_type}")
async def get_report(query: str, report_type: str) -> dict:
return {"report": query, "type": report_type}
```
Output: {"report":"What is 42?","type":"outline_report"}
URL2 is used for the GPT-Researcher FastAPI call test which is resulting in the error.
```
@app.get("/api/report/{query}/{report_type}")
async def get_report(query: str, report_type: str) -> dict:
researcher = GPTResearcher(query, report_type)
research_result = await researcher.conduct_research()
report = await researcher.write_report()
return {"report": report}
```
It appears to be a utf-8 issue perhaps,
However, I don't encounter this on Windows when running the actual gpt-researcher code.
ref: https://ask.replit.com/t/unicodeencodeerror-charmap-codec-cant-encode-character-ufb01/76667
Then again, running this as an api call is a little different from the regular FE/BE GPT-R.
Any clues?
Full error log is:
```
] INFO: 127.0.0.1:52833 - "GET /api/reporttest/What%20is%2042%3F/outline_report HTTP/1.1" 200 OK
[0] GET / 200 in 28ms
[1] INFO: 127.0.0.1:52843 - "GET /api/report/What%20is%2042%3F/outline_report HTTP/1.1" 500 Internal Server Error
[1] ERROR: Exception in ASGI application
[1] Traceback (most recent call last):
[1] File "E:\Source Control\AI Apps\Python\nextjs-fastapi\.venv\Lib\site-packages\uvicorn\protocols\http\httptools_impl.py", line 399, in run_asgi
[1] result = await app( # type: ignore[func-returns-value]
[1] ^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
[1] File "E:\Source Control\AI Apps\Python\nextjs-fastapi\.venv\Lib\site-packages\uvicorn\middleware\proxy_headers.py", line 70, in __call__
[1] return await self.app(scope, receive, send)
[1] ^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
[1] File "E:\Source Control\AI Apps\Python\nextjs-fastapi\.venv\Lib\site-packages\fastapi\applications.py", line 1054, in __call__
[1] await super().__call__(scope, receive, send)
[1] File "E:\Source Control\AI Apps\Python\nextjs-fastapi\.venv\Lib\site-packages\starlette\applications.py", line 123, in __call__
[1] await self.middleware_stack(scope, receive, send)
[1] File "E:\Source Control\AI Apps\Python\nextjs-fastapi\.venv\Lib\site-packages\starlette\middleware\errors.py", line 186, in __call__
[1] raise exc
[1] File "E:\Source Control\AI Apps\Python\nextjs-fastapi\.venv\Lib\site-packages\starlette\middleware\errors.py", line 164, in __call__
[1] await self.app(scope, receive, _send)
[1] File "E:\Source Control\AI Apps\Python\nextjs-fastapi\.venv\Lib\site-packages\starlette\middleware\exceptions.py", line 65, in __call__
[1] await wrap_app_handling_exceptions(self.app, conn)(scope, receive, send)
[1] File "E:\Source Control\AI Apps\Python\nextjs-fastapi\.venv\Lib\site-packages\starlette\_exception_handler.py", line 64, in wrapped_app
[1] raise exc
[1] File "E:\Source Control\AI Apps\Python\nextjs-fastapi\.venv\Lib\site-packages\starlette\_exception_handler.py", line 53, in wrapped_app
[1] await app(scope, receive, sender)
[1] File "E:\Source Control\AI Apps\Python\nextjs-fastapi\.venv\Lib\site-packages\starlette\routing.py", line 756, in __call__
[1] await self.middleware_stack(scope, receive, send)
[1] File "E:\Source Control\AI Apps\Python\nextjs-fastapi\.venv\Lib\site-packages\starlette\routing.py", line 776, in app
[1] await route.handle(scope, receive, send)
[1] File "E:\Source Control\AI Apps\Python\nextjs-fastapi\.venv\Lib\site-packages\starlette\routing.py", line 297, in handle
[1] await self.app(scope, receive, send)
[1] File "E:\Source Control\AI Apps\Python\nextjs-fastapi\.venv\Lib\site-packages\starlette\routing.py", line 77, in app
[1] await wrap_app_handling_exceptions(app, request)(scope, receive, send)
[1] File "E:\Source Control\AI Apps\Python\nextjs-fastapi\.venv\Lib\site-packages\starlette\_exception_handler.py", line 64, in wrapped_app
[1] raise exc
[1] File "E:\Source Control\AI Apps\Python\nextjs-fastapi\.venv\Lib\site-packages\starlette\_exception_handler.py", line 53, in wrapped_app
[1] await app(scope, receive, sender)
[1] File "E:\Source Control\AI Apps\Python\nextjs-fastapi\.venv\Lib\site-packages\starlette\routing.py", line 72, in app
[1] response = await func(request)
[1] ^^^^^^^^^^^^^^^^^^^
[1] File "E:\Source Control\AI Apps\Python\nextjs-fastapi\.venv\Lib\site-packages\fastapi\routing.py", line 278, in app
[1] raw_response = await run_endpoint_function(
[1] ^^^^^^^^^^^^^^^^^^^^^^^^^^^^
[1] File "E:\Source Control\AI Apps\Python\nextjs-fastapi\.venv\Lib\site-packages\fastapi\routing.py", line 191, in run_endpoint_function
[1] return await dependant.call(**values)
[1] ^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
[1] File "E:\Source Control\AI Apps\Python\nextjs-fastapi\api\index.py", line 23, in get_report
[1] research_result = await researcher.conduct_research()
[1] ^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
[1] File "E:\Source Control\AI Apps\Python\nextjs-fastapi\.venv\Lib\site-packages\gpt_researcher\master\agent.py", line 77, in conduct_research
[1] await stream_output("logs", f"\U0001f50e Starting the research task for '{self.query}'...", self.websocket)
[1] File "E:\Source Control\AI Apps\Python\nextjs-fastapi\.venv\Lib\site-packages\gpt_researcher\master\actions.py", line 321, in stream_output
[1] print(output)
[1] File "D:\Python\Lib\encodings\cp1252.py", line 19, in encode
[1] return codecs.charmap_encode(input,self.errors,encoding_table)[0]
[1] ^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
[1] UnicodeEncodeError: 'charmap' codec can't encode character '\U0001f50e' in position 0: character maps to <undefined>
```
|
closed
|
2024-07-06T05:39:07Z
|
2024-07-07T03:38:54Z
|
https://github.com/assafelovic/gpt-researcher/issues/645
|
[] |
sonicviz
| 5
|
gunthercox/ChatterBot
|
machine-learning
| 2,033
|
Creating a chatbot
|
Errors in importing the chatterbot and installing Chatbot to python
|
closed
|
2020-08-27T15:48:33Z
|
2025-02-26T11:43:08Z
|
https://github.com/gunthercox/ChatterBot/issues/2033
|
[] |
Anwarite
| 4
|
netbox-community/netbox
|
django
| 18,327
|
error after update to NetBox 4.2.0: requires_internet
|
### Deployment Type
Self-hosted
### Triage priority
N/A
### NetBox Version
v4.2.0
### Python Version
3.12
### Steps to Reproduce
After updating NetBox to 4.20 using the Git method. Login Page works fine if not logged in. After login the error occurs.
### Expected Behavior
Can Login normally.
### Observed Behavior
Server Error
There was a problem with your request. Please contact an administrator.
The complete exception is provided below:
<class 'KeyError'>
'requires_internet'
Python version: 3.12.3
NetBox version: 4.2.0
Plugins: None installed
If further assistance is required, please post to the [NetBox discussion forum](https://github.com/netbox-community/netbox/discussions) on GitHub.
|
closed
|
2025-01-07T15:13:52Z
|
2025-01-07T15:36:49Z
|
https://github.com/netbox-community/netbox/issues/18327
|
[
"type: bug",
"status: duplicate"
] |
Kujo01243
| 4
|
babysor/MockingBird
|
deep-learning
| 794
|
PPG训练时的报错,请帮忙看看
|
PPG预处理很顺利,ppg2mel.yaml路径也改了,但是这个错误提示怎么都解决不了,请大家帮忙看看能否有经验分享下。
D:\MockingBird-main>python ppg2mel_train.py --config .\ppg2mel\saved_models\ppg2mel.yaml --oneshotvc
Traceback (most recent call last):
File "D:\MockingBird-main\ppg2mel_train.py", line 67, in <module>
main()
File "D:\MockingBird-main\ppg2mel_train.py", line 50, in main
config = HpsYaml(paras.config)
File "D:\MockingBird-main\utils\load_yaml.py", line 44, in __init__
hps = load_hparams(yaml_file)
File "D:\MockingBird-main\utils\load_yaml.py", line 8, in load_hparams
for doc in docs:
File "C:\Users\benny\AppData\Local\Programs\Python\Python39\lib\site-packages\yaml\__init__.py", line 127, in load_all
loader = Loader(stream)
File "C:\Users\benny\AppData\Local\Programs\Python\Python39\lib\site-packages\yaml\loader.py", line 34, in __init__
Reader.__init__(self, stream)
File "C:\Users\benny\AppData\Local\Programs\Python\Python39\lib\site-packages\yaml\reader.py", line 85, in __init__
self.determine_encoding()
File "C:\Users\benny\AppData\Local\Programs\Python\Python39\lib\site-packages\yaml\reader.py", line 124, in determine_encoding
self.update_raw()
File "C:\Users\benny\AppData\Local\Programs\Python\Python39\lib\site-packages\yaml\reader.py", line 178, in update_raw
data = self.stream.read(size)
UnicodeDecodeError: 'gbk' codec can't decode byte 0x86 in position 176: illegal multibyte sequence
|
open
|
2022-12-01T14:03:43Z
|
2022-12-03T02:45:27Z
|
https://github.com/babysor/MockingBird/issues/794
|
[] |
benny1227
| 1
|
psf/requests
|
python
| 6,830
|
PreparedRequests can't bypass URL normalization when proxies are used
|
Related to #5289, where [akmalhisyam found a way to bypass URL normalization using PreparedRequests](https://github.com/psf/requests/issues/5289#issuecomment-573632625), however, the solution doesn't work when you have proxies provided.
## Expected Result
This should be able to explicitly set the request URL without getting normalized (from `/../something.txt` to `/something.txt`)
```
url = "http://example.com/../something.txt"
s = requests.Session()
req = requests.Request(method='POST' ,url=url, headers=headers, data=data)
prep = req.prepare()
prep.url = url
r = s.send(prep, proxies={"http": "http://127.0.0.1"}, verify=False)
```
## Actual Result
The code above doesn't work, this one works though:
```
url = "http://example.com/../something.txt"
s = requests.Session()
req = requests.Request(method='POST' ,url=url, headers=headers, data=data)
prep = req.prepare()
prep.url = url
r = s.send(prep, verify=False)
```
## Reproduction Steps
Use the code in **Expected Result** and check your proxy request log, you will see it doesn't work
## System Information
$ python -m requests.help
```json
{
"chardet": {
"version": "5.2.0"
},
"charset_normalizer": {
"version": "2.0.12"
},
"cryptography": {
"version": "38.0.4"
},
"idna": {
"version": "3.4"
},
"implementation": {
"name": "CPython",
"version": "3.11.4"
},
"platform": {
"release": "4.4.0-19041-Microsoft",
"system": "Linux"
},
"pyOpenSSL": {
"openssl_version": "30000080",
"version": "21.0.0"
},
"requests": {
"version": "2.32.3"
},
"system_ssl": {
"version": "30000030"
},
"urllib3": {
"version": "2.0.4"
},
"using_charset_normalizer": false,
"using_pyopenssl": true
}
```
|
open
|
2024-11-18T17:06:17Z
|
2025-01-27T18:44:36Z
|
https://github.com/psf/requests/issues/6830
|
[] |
shelld3v
| 1
|
microsoft/nlp-recipes
|
nlp
| 285
|
[ASK] Add ReadMe for subfolder unit under tests
|
### Description
Add a ReadaMe file descriping the scope of all unit tests. Are we having full coverage of units tests for all utils and notebooks?
### Other Comments
**Principles of NLP Documentation**
Each landing page at the folder level should have a ReadMe which explains -
○ Summary of what this folder offers.
○ Why and how it benefits users
○ As applicable - Documentation of using it, brief description etc
**Scenarios folder:**
○ Root Scenario folder should have a summary on what value these example notebook provides.
○ Include a table with scenario name, description, algorithm, Dataset
○ Other instructions, Pre-req of running these notebooks
○ Each scenario folder should have a summary text explaining about the scenario, what utils its using. Any benchmark numbers if applicable. Explain any concept relevant to the scenario
○ Under each scenario folder there should be one Quick Start example notebook, name starting with "QuickStart: ..." and atleast one AML notebook
**Example Notebooks Guiding Principles:**
○ We are providing recipes for solving NLP scenarios on Azure AI
○ We make it easier by providing Util packages
○ We provide example notebooks on how to use the utils for solving common NLP scenarios
○ Based on these principles above, all notebook examples should be using utils wherever applicable. Ex: If your example is doing classification using BERT, use the BERTSequenceClassifier instead of directly calling BertForSequenceClassification. Same with tokenization.
|
closed
|
2019-08-13T22:13:37Z
|
2019-08-16T20:56:48Z
|
https://github.com/microsoft/nlp-recipes/issues/285
|
[
"documentation",
"release-blocker"
] |
dipanjan77
| 1
|
globaleaks/globaleaks-whistleblowing-software
|
sqlalchemy
| 4,345
|
Verify possibility to restrict the content security policy preventing usage of inline CSS styles
|
### Proposal
This ticket is to keep track of the activities related to verify possibility to restrict the content security policy preventing usage of inline styles and the possible implementations necessary to achieve this goal on globaleaks v5 as previously achieved on client globaleaks 4.
At the moment it seems that the library requiring to used inline styles is only: [ng-bootstrap](https://github.com/ng-bootstrap/ng-bootstrap/issues/2085) that uses inline styles for example for the tooltip and calendar implementations.
|
closed
|
2024-12-03T13:44:20Z
|
2024-12-06T16:14:48Z
|
https://github.com/globaleaks/globaleaks-whistleblowing-software/issues/4345
|
[
"T: Enhancement",
"C: Client",
"C: Backend",
"F: Security"
] |
evilaliv3
| 1
|
bigscience-workshop/petals
|
nlp
| 282
|
http://health.petals.ml/ shows "broken" in the same timeframe i spin up a docker petals
|
Hi there.
I think your project is awesome and want to support you by sharing my resources.
I noticed an outage on bloom and bloomz, yesterday when i used a dockercompose to spin up a new docker image.
I noticed a second outage just now on bloom, when i started the colab on https://colab.research.google.com/drive/1Ervk6HPNS6AYVr3xVdQnY5a-TjjmLCdQ?usp=sharing
Maybe its just coincidence, but maybe some script for taking the new node in crashes your config something...?
Just let you know in case.
Thanks for that awesome project.
|
closed
|
2023-03-09T12:15:15Z
|
2023-03-09T23:59:01Z
|
https://github.com/bigscience-workshop/petals/issues/282
|
[] |
worldpeaceenginelabs
| 1
|
biolab/orange3
|
pandas
| 7,009
|
Metavariables are not excluded in feature selection methods
|
A longstanding issue is that metavariables are not excluded from methods. For example, in "find informative projections" for scatter plots, they appear as suggestions. Also, in feature suggestions, the metas are included. If there are many, the automatic feature selection breaks down. This is a nuisance, as metas often contain the solution to a classification problem. "Find informative mosaics" has the same issue, as does the violin plot where ordering by relevance also includes metas. Tree prediction does ignore them, though.
I am currently using version 338 on a Mac, and this error is present in the PC version as well.
This issue has existed in every version of Orange that I can recall.
Best larerooreal
|
open
|
2025-01-30T13:21:18Z
|
2025-02-19T10:01:28Z
|
https://github.com/biolab/orange3/issues/7009
|
[
"needs discussion",
"bug report"
] |
lareooreal
| 4
|
scrapy/scrapy
|
python
| 5,944
|
Improve statscollector.py along with test_stats.py and test_link.py
|
## Summary
Remove unused code from statscollector.py and improve test suits in test_stats.py and test_link.py
## Motivation
I was working with the project and reviewed some of the code trying to understand how the stats collection is working and I noticed that some of the code hasn't been implemented in the statscollectors.py file as well as the testing suites in test_stats.py and test_link.py are not properly testing the files. I wanted to bring up these issues to improve the project, increase the testing coverage for statscollector.py and make it easier to maintain.
## Describe alternatives you've considered
Reviewing the files, I think the best approach is to remove the DummyStatsCollector class in the statscollector.py file since it is not used anywhere in the project and complete the open_spider and _persist_stats methods in the StatsCollector class. In the test_stats.py and test_link.py files, the best approach would be to separate the tests in unique test cases and fill in for the missing methods in the test_stats.py file in order to increase testing coverage.
|
closed
|
2023-06-04T20:58:39Z
|
2023-06-21T09:20:59Z
|
https://github.com/scrapy/scrapy/issues/5944
|
[] |
DeanDro
| 1
|
chatopera/Synonyms
|
nlp
| 56
|
请问下相似度计算公式是什么?
|
请问下相似度计算公式是什么?
目前我用的多的是textrank + word2vec
请问本工具的算法是?我想做下对比,可能的话我把我的算法也pr过来
|
closed
|
2018-03-19T15:48:23Z
|
2018-03-24T23:05:03Z
|
https://github.com/chatopera/Synonyms/issues/56
|
[] |
Wall-ee
| 2
|
wkentaro/labelme
|
computer-vision
| 459
|
Add shortcut for ‘Add Point to Edge’
|
Hello, i really appreciate your work, the software really helps me a lot.
I want to add shortcut for ‘Add Point to Edge’, but it does not work. The mouse needs to be in a specific location, how can I add shortcut?
|
closed
|
2019-08-07T11:10:25Z
|
2019-08-23T10:00:40Z
|
https://github.com/wkentaro/labelme/issues/459
|
[] |
stormlands
| 2
|
mkhorasani/Streamlit-Authenticator
|
streamlit
| 71
|
Saving cookie failed when deploying the app with docker
|
Hey everybody,
I implemented the auth as described in your README and tested it locally on my machine - works fine. Then i deployed the same app using Docker and the authentification does not work as expected. Seems like the client-side cookies are not saved when using Docker.
Does anyone know the problem or even the solution?
Thanks a lot!
|
open
|
2023-06-12T07:23:31Z
|
2024-07-27T14:42:48Z
|
https://github.com/mkhorasani/Streamlit-Authenticator/issues/71
|
[
"help wanted"
] |
ArnoSchiller
| 3
|
QingdaoU/OnlineJudge
|
django
| 50
|
2.0重构版规划
|
本OJ大约是1年前开始开发的,目前逐渐暴露出一些问题。打算进行一个比较大的重构,主要包括
- [x] vue.js重写所有前端页面
- [x] 前端后端的国际化,多语言和时区
- [x] 导入导出题目(但是目前考虑的hustoj的FPS格式还有些问题,可能在当前版本中也会做)
- [x] 更方便的添加多编程程序语言支持,统一配置规则(有些问题还没解决)题目选择可以使用的语言
- [x] 比赛的OI模式(排名,查看单独测试数据是否通过等)
- [x] SPJ更加方便的添加代码和测试
- [x] 超级管理员 - 管理所有 普通管理员- 默认创建小组内比赛和不能创建题目题目,但是可以通过两个选项允许
- [x] 完善后端的测试,跑CI
- [x] 代码风格问题
- [x] 题目和公告使用markdown编辑器
- [x] 判题服务器的健康检查和添加时候的检查
目前基本上只有我一个人在维护这个项目,时间不多,都是在工作之余做,但是放弃了也太可惜。如果有愿意参与的,可以回复一下。
扫描二维码献出你的一份爱心

|
closed
|
2016-06-24T05:32:44Z
|
2019-01-05T06:15:41Z
|
https://github.com/QingdaoU/OnlineJudge/issues/50
|
[] |
virusdefender
| 27
|
JaidedAI/EasyOCR
|
pytorch
| 1,335
|
FileNotFoundError in `download_and_unzip` when running multiple easyocr's concurrently
|
When we try to run two or more easyocr's concurrently, we get an error in the downloader. I am guessing that the download logic uses a fixed download filepath?
```shell
EasyOcrModel(
File ".../lib/python3.10/site-packages/docling/models self.reader = easyocr.Reader(config["lang"])
File ".../lib/python3.10/site-packages/easyocr/easyocr.py", line 92, in __init__
detector_path = self.getDetectorPath(detect_network)
File ".../lib/python3.10/site-packages/easyocr/easyocr.py", line 253, in getDetectorPath
download_and_unzip(self.detection_models[self.detect_network]['url'], self.detection_models[self.detect_network]['filename'], self.model_storage_directory, self.verbose)
File ".../lib/python3.10/site-packages/easyocr/utils.py", line 631, in download_and_unzip
os.remove(zip_path)
FileNotFoundError: [Errno 2] No such file or directory: '/home/runner/.EasyOCR//model/temp.zip'
```
|
open
|
2024-11-18T21:39:58Z
|
2024-12-18T10:27:48Z
|
https://github.com/JaidedAI/EasyOCR/issues/1335
|
[] |
starpit
| 2
|
tflearn/tflearn
|
tensorflow
| 960
|
TypeError: only integer scalar arrays can be converted to a scalar index
|
Exception in thread Thread-8:
Traceback (most recent call last):
File "C:\Users\Bhumit\Anaconda3\lib\threading.py", line 916, in _bootstrap_inner
self.run()
File "C:\Users\Bhumit\Anaconda3\lib\threading.py", line 864, in run
self._target(*self._args, **self._kwargs)
File "C:\Users\Bhumit\Anaconda3\lib\site-packages\tflearn\data_flow.py", line 187, in fill_feed_dict_queue
data = self.retrieve_data(batch_ids)
File "C:\Users\Bhumit\Anaconda3\lib\site-packages\tflearn\data_flow.py", line 222, in retrieve_data
utils.slice_array(self.feed_dict[key], batch_ids)
File "C:\Users\Bhumit\Anaconda3\lib\site-packages\tflearn\utils.py", line 187, in slice_array
return X[start]
TypeError: only integer scalar arrays can be converted to a scalar index
Anyone, please suggest solutions for above errors
|
open
|
2017-11-17T13:02:41Z
|
2017-11-17T13:02:41Z
|
https://github.com/tflearn/tflearn/issues/960
|
[] |
AdivarekarBhumit
| 0
|
blb-ventures/strawberry-django-plus
|
graphql
| 256
|
Using input_mutation with a None return type throws an exception
|
I have a mutation that looks something like
```python
@gql.django.input_mutation(permission_classes=[IsAdmin])
def mutate_thing(
self,
info: Info,
) -> None:
# do the thing
return None
```
This throws an exception when I try to generate my schema:
```
File "/Users/tao/dev/cinder/myapp/mutations.py", line 599, in Mutation
@gql.django.input_mutation(permission_classes=[IsAdmin])
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/Users/tao/dev/cinder/.venv/lib/python3.11/site-packages/strawberry_django_plus/mutations/fields.py", line 126, in __call__
types_ = tuple(get_possible_types(annotation.resolve()))
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/Users/tao/dev/cinder/.venv/lib/python3.11/site-packages/strawberry_django_plus/utils/inspect.py", line 171, in get_possible_types
assert_never(gql_type)
File "/Users/tao/.asdf/installs/python/3.11.1/lib/python3.11/typing.py", line 2459, in assert_never
raise AssertionError(f"Expected code to be unreachable, but got: {value}")
AssertionError: Expected code to be unreachable, but got: None
```
It looks like the issue occurs in `get_possible_types`, which doesn't handle a None return type. It's possible to work around this by setting the return type annotation to `Void._scalar_definition` instead of `None`, but that feels like a hack!
|
open
|
2023-07-02T21:39:36Z
|
2023-07-03T13:15:57Z
|
https://github.com/blb-ventures/strawberry-django-plus/issues/256
|
[] |
taobojlen
| 1
|
DistrictDataLabs/yellowbrick
|
matplotlib
| 560
|
Add 'Yellowbrick for Teachers' slidedeck to docs
|
**Describe the solution you'd like**
Add a sample slide deck to the docs for machine learning teachers to use in teaching model selection & visual diagnostics
**Is your feature request related to a problem? Please describe.**
I've been asked a few times by other teachers of machine learning if I had any slides or teaching materials they could use in their classes. I have some slides that I could put together and publish, or else this might be a good candidate for [Jupyter Notebook slides](https://medium.com/@mjspeck/presenting-code-using-jupyter-notebook-slides-a8a3c3b59d67)
**Examples**
<img width="801" alt="screen shot 2018-08-10 at 2 36 48 pm" src="https://user-images.githubusercontent.com/8760385/43975216-dd9a8d14-9caa-11e8-9f7d-2466cd58df44.png">
<img width="802" alt="screen shot 2018-08-10 at 2 37 31 pm" src="https://user-images.githubusercontent.com/8760385/43975245-f456554c-9caa-11e8-91ef-1b8d1263c05b.png">
|
closed
|
2018-08-10T18:38:19Z
|
2018-08-13T15:17:14Z
|
https://github.com/DistrictDataLabs/yellowbrick/issues/560
|
[] |
rebeccabilbro
| 3
|
gee-community/geemap
|
jupyter
| 2,011
|
error with 'geemap.requireJS'
|
<!-- Please search existing issues to avoid creating duplicates. -->
### Environment Information

### Description
error with 'geemap.requireJS'
### What I Did
```
import geemap
WS = geemap.requireJS('users/dushuai/showANDdownload_rec_of_rgb:learningCode_from_articles/whittaker_smoother')
---------------------------------------------------------------------------
ModuleNotFoundError Traceback (most recent call last)
File E:\ProgramData\Anaconda3\envs\gee1\Lib\site-packages\geemap\common.py:13482, in requireJS(lib_path, Map)
13481 try:
> 13482 from oeel import oeel
13483 except ImportError:
File E:\ProgramData\Anaconda3\envs\gee1\Lib\site-packages\oeel\oeel.py:17
16 from . import external
---> 17 from . import colab
18 oeelLibPath=os.path.dirname(__file__)
File E:\ProgramData\Anaconda3\envs\gee1\Lib\site-packages\oeel\colab.py:3
2 import IPython
----> 3 from google.colab import output
4 from google.oauth2.credentials import Credentials
File E:\ProgramData\Anaconda3\envs\gee1\Lib\site-packages\google\colab\__init__.py:23
22 from google.colab import _shell_customizations
---> 23 from google.colab import _system_commands
24 from google.colab import _tensorflow_magics
File E:\ProgramData\Anaconda3\envs\gee1\Lib\site-packages\google\colab\_system_commands.py:24
23 import os
---> 24 import pty
25 import select
File E:\ProgramData\Anaconda3\envs\gee1\Lib\pty.py:12
11 import sys
---> 12 import tty
14 # names imported directly for test mocking purposes
File E:\ProgramData\Anaconda3\envs\gee1\Lib\tty.py:5
3 # Author: Steen Lumholt.
----> 5 from termios import *
7 __all__ = ["setraw", "setcbreak"]
ModuleNotFoundError: No module named 'termios'
During handling of the above exception, another exception occurred:
ImportError Traceback (most recent call last)
Cell In[7], line 2
1 import geemap
----> 2 WS = geemap.requireJS('users/dushuai/showANDdownload_rec_of_rgb:learningCode_from_articles/whittaker_smoother')
File E:\ProgramData\Anaconda3\envs\gee1\Lib\site-packages\geemap\common.py:13484, in requireJS(lib_path, Map)
13482 from oeel import oeel
13483 except ImportError:
> 13484 raise ImportError(
13485 "oeel is required for requireJS. Please install it using 'pip install oeel'."
13486 )
13488 ee_initialize()
13490 if lib_path is None:
ImportError: oeel is required for requireJS. Please install it using 'pip install oeel'.
```
|
closed
|
2024-05-14T02:45:35Z
|
2024-05-16T18:05:29Z
|
https://github.com/gee-community/geemap/issues/2011
|
[
"bug"
] |
Dushuai12138
| 5
|
liangliangyy/DjangoBlog
|
django
| 191
|
Centos 7 + apache2 部署
|
请问这是什么错误啊...弄了一天实在没头绪
|
closed
|
2018-12-08T09:11:07Z
|
2018-12-11T10:22:49Z
|
https://github.com/liangliangyy/DjangoBlog/issues/191
|
[] |
FishWoWater
| 5
|
Evil0ctal/Douyin_TikTok_Download_API
|
fastapi
| 166
|
[BUG] Tiktok接口是挂了吗?
|
最近发现接口频繁返回:{"status_code":0,"status_msg":"","block_code":2018},貌似现在强制性需要提供X-Argus、X-Ladon两个算参数才能返回了,但我发现douyin.wtf的接口是可以正常拿到数据的,有点疑惑,难道是因为ip的问题?
|
closed
|
2023-03-08T02:24:42Z
|
2023-03-09T02:35:00Z
|
https://github.com/Evil0ctal/Douyin_TikTok_Download_API/issues/166
|
[
"BUG"
] |
juedi998
| 5
|
pydantic/pydantic-ai
|
pydantic
| 496
|
Configuration and parameters for `all_messages()` and `new_messages()`
|
It would be helpful if the `all_messages()` and `new_messages()` methods had an option to exclude the system prompt like `all_messages(system_prompt=False)`. This would probably be a better default behavior too. Why?
Well, when do you use these methods?
### 1. Passing messages to the client/frontend
```python
@app.post("/chat")
async def chat(data):
...
result = await agent.run(data.message, message_history=data.history)
return result.all_messages()
```
You probably don't want to pass the system prompt along.
### 2. When storing messages in a database
```python
...
result = await agent.run(data.message, message_history=data.history)
db.table("conversations").insert(result.all_messages_json())
...
```
You probably don't want to store the system prompt for every conversation.
### 3. When handing over a conversation to a different agent
```python
result1 = agent1.run(message, message_history=history)
...
# handover detected
result2 = agent2.run(message, message_history=result1.all_messages())
````
You want the chat messages for context, but the system prompt of the new agent.
---
More parameters to exclude tool calls or just tool call responses would be another great addition, I think.
|
open
|
2024-12-19T12:06:26Z
|
2025-02-17T04:24:12Z
|
https://github.com/pydantic/pydantic-ai/issues/496
|
[
"Feature request"
] |
pietz
| 7
|
amidaware/tacticalrmm
|
django
| 1,913
|
tactical meshagent memory leak
|
**Server Info (please complete the following information):**
- OS: Ubuntu 22.04.4
- Browser: chrome
- RMM Version (as shown in top left of web UI): v0.18.2
**Installation Method:**
- [ x ] Standard
**Agent Info (please complete the following information):**
- Agent version (as shown in the 'Summary' tab of the agent from web UI): Agent v2.7.0
- Agent OS: Ubuntu 22.04.4
**Describe the bug**
tactical meshagent service has a memory leak and slowly grows to 10s of gigabytes of memory usage until it is restarted.
**To Reproduce**
Steps to reproduce the behavior:
1. install tactical agent on ubuntu 22.04.4
**Expected behavior**
agent uses a reasonable amount of ram and doesnt need to be restarted to reduce memory usage
**Screenshots**

|
closed
|
2024-07-08T21:54:19Z
|
2024-07-10T05:10:02Z
|
https://github.com/amidaware/tacticalrmm/issues/1913
|
[] |
slapplebags
| 1
|
MaxHalford/prince
|
scikit-learn
| 144
|
Not compatible with pandas 2.0.0
|
I'm having dependency conflicts when trying to install prince and pandas==2.0.0 in the same environment.
|
closed
|
2023-04-17T20:42:03Z
|
2023-04-18T12:56:10Z
|
https://github.com/MaxHalford/prince/issues/144
|
[] |
JuanCruzC97
| 4
|
scikit-learn/scikit-learn
|
python
| 30,699
|
Make scikit-learn OpenML more generic for the data download URL
|
According to https://github.com/orgs/openml/discussions/20#discussioncomment-11913122 our code hardcodes where to find the OpenML data.
I am not quite sure what needs to be done right now but maybe @PGijsbers has some suggestions (not urgent at all though, I am guessing you have bigger fish to fry right now 😉) or maybe @glemaitre .
|
closed
|
2025-01-22T09:13:44Z
|
2025-02-25T15:09:52Z
|
https://github.com/scikit-learn/scikit-learn/issues/30699
|
[
"Enhancement",
"module:datasets"
] |
lesteve
| 3
|
oegedijk/explainerdashboard
|
dash
| 147
|
Add support to Imbalanced-learn pipelines in ClassifierExplainer
|
When I try to generate a `ClassifierExplainer``on a imblearn pipeline I get the following **error**:
```
TypeError: All intermediate steps should be transformers and implement fit and transform or be the string 'passthrough'
'SMOTETomek(random_state=42)' (type <class 'imblearn.combine._smote_tomek.SMOTETomek'>) doesn't
```
**Full traceback:**
```
---------------------------------------------------------------------------
TypeError Traceback (most recent call last)
<ipython-input-75-2a92cf12a19d> in <module>
1 from explainerdashboard import ClassifierExplainer, InlineExplainer
----> 2 explainer = ClassifierExplainer(best_model, X_test, y_test)
/opt/conda/lib/python3.7/site-packages/explainerdashboard/explainers.py in __init__(self, model, X, y, permutation_metric, shap, X_background, model_output, cats, cats_notencoded, idxs, index_name, target, descriptions, n_jobs, permutation_cv, cv, na_fill, precision, labels, pos_label)
1999 cats, cats_notencoded, idxs, index_name, target,
2000 descriptions, n_jobs, permutation_cv, cv, na_fill,
-> 2001 precision)
2002
2003 assert hasattr(model, "predict_proba"), \
/opt/conda/lib/python3.7/site-packages/explainerdashboard/explainers.py in __init__(self, model, X, y, permutation_metric, shap, X_background, model_output, cats, cats_notencoded, idxs, index_name, target, descriptions, n_jobs, permutation_cv, cv, na_fill, precision)
138 if shap != 'kernel':
139 pipeline_model = model.steps[-1][1]
--> 140 pipeline_transformer = Pipeline(model.steps[:-1])
141 if hasattr(model, "predict") and hasattr(pipeline_transformer, "transform"):
142 X_transformed = pipeline_transformer.transform(X)
/opt/conda/lib/python3.7/site-packages/sklearn/utils/validation.py in inner_f(*args, **kwargs)
61 extra_args = len(args) - len(all_args)
62 if extra_args <= 0:
---> 63 return f(*args, **kwargs)
64
65 # extra_args > 0
/opt/conda/lib/python3.7/site-packages/sklearn/pipeline.py in __init__(self, steps, memory, verbose)
116 self.memory = memory
117 self.verbose = verbose
--> 118 self._validate_steps()
119
120 def get_params(self, deep=True):
/opt/conda/lib/python3.7/site-packages/sklearn/pipeline.py in _validate_steps(self)
169 "transformers and implement fit and transform "
170 "or be the string 'passthrough' "
--> 171 "'%s' (type %s) doesn't" % (t, type(t)))
172
173 # We allow last estimator to be None as an identity transformation
TypeError: All intermediate steps should be transformers and implement fit and transform or be the string 'passthrough' 'SMOTETomek(random_state=42)' (type <class 'imblearn.combine._smote_tomek.SMOTETomek'>) doesn't
```
**Versions**
System:
python: 3.7.10 | packaged by conda-forge | (default, Feb 19 2021, 16:07:37) [GCC 9.3.0]
executable: /opt/conda/bin/python
machine: Linux-5.4.120+-x86_64-with-debian-buster-sid
Python dependencies:
pip: 21.1.2
setuptools: 49.6.0.post20210108
sklearn: 0.24.2
imblearn: 0.8.0
explainerdashboard: latest
numpy: 1.19.5
scipy: 1.6.3
Cython: 0.29.23
pandas: 1.2.4
matplotlib: 3.4.2
joblib: 1.0.1
threadpoolctl: 2.1.0
|
closed
|
2021-09-17T12:18:59Z
|
2021-12-26T21:25:17Z
|
https://github.com/oegedijk/explainerdashboard/issues/147
|
[] |
Abdelgha-4
| 6
|
jowilf/starlette-admin
|
sqlalchemy
| 563
|
Bug: sorting by datetime does not work
|
hi
postgres+asyncpg
starlette-admin==0.13.2
models.py
```python
class Log(Base):
created_at: Mapped[datetime] = mapped_column(DateTime(timezone=True), server_default=func.now())
...
```
```
2024-07-17 12:25:35.227 UTC [4632] ERROR: operator does not exist: timestamp with time zone >= character varying at character 156
2024-07-17 12:25:35.227 UTC [4632] HINT: No operator matches the given name and argument types. You might need to add explicit type casts.
2024-07-17 12:25:35.227 UTC [4632] STATEMENT: SELECT logs.url, logs.endpoint, logs.created_at, logs.status_code, logs.content_length, logs.api_key, logs.body, logs.id
FROM logs
WHERE logs.created_at BETWEEN $1::VARCHAR AND $2::VARCHAR ORDER BY logs.created_at DESC
LIMIT $3::INTEGER OFFSET $4::INTEGER
(sqlalchemy.dialects.postgresql.asyncpg.ProgrammingError) <class 'asyncpg.exceptions.UndefinedFunctionError'>: operator does not exist: timestamp with time zone >= character varying
HINT: No operator matches the given name and argument types. You might need to add explicit type casts.
[SQL: SELECT logs.url, logs.endpoint, logs.created_at, logs.status_code, logs.content_length, logs.api_key, logs.body, logs.id
FROM logs
WHERE logs.created_at BETWEEN $1::VARCHAR AND $2::VARCHAR ORDER BY logs.created_at DESC
LIMIT $3::INTEGER OFFSET $4::INTEGER]
[parameters: ('2024-07-17T15:25:00+03:00', '2024-07-18T15:25:00+03:00', 100, 0)]
(Background on this error at: https://sqlalche.me/e/20/f405)
2024-07-17 12:25:35.229 UTC [4632] ERROR: current transaction is aborted, commands ignored until end of transaction block
2024-07-17 12:25:35.229 UTC [4632] STATEMENT: SELECT logs.url, logs.endpoint, logs.created_at, logs.status_code, logs.content_length, logs.api_key, logs.body, logs.id
FROM logs
WHERE logs.created_at BETWEEN $1::VARCHAR AND $2::VARCHAR ORDER BY logs.created_at DESC
LIMIT $3::INTEGER OFFSET $4::INTEGER
INFO: 172.21.0.1:51892 - "GET /admin/api/log?skip=0&limit=100&order_by=created_at%20desc&where=%7B%22and%22%3A%5B%7B%22created_at%22%3A%7B%22between%22%3A%5B%222024-07-17T15%3A25%3A00%2B03%3A00%22%2C%222024-07-18T15%3A25%3A00%2B03%3A00%22%5D%7D%7D%5D%7D HTTP/1.1" 500 Internal Server Error
```
how do I fix it?
|
open
|
2024-07-17T12:29:13Z
|
2025-03-19T14:43:56Z
|
https://github.com/jowilf/starlette-admin/issues/563
|
[
"bug"
] |
Kaiden0001
| 1
|
youfou/wxpy
|
api
| 46
|
如何根据接收的消息在多线程中控制注册消息?
|
文档中说明要在额外的线程控制开关注册消息。
但我在代码中测试过程中,发现在单个线程中也能个进行开关注册。
想问问作者多线程中如何实现,谢谢
|
closed
|
2017-05-04T09:37:39Z
|
2017-05-06T08:20:56Z
|
https://github.com/youfou/wxpy/issues/46
|
[
"question"
] |
cxyfreedom
| 1
|
coqui-ai/TTS
|
pytorch
| 3,142
|
Fairseq voice cloning
|
### Describe the bug
There seems to be an issue of activating voice conversion in Coqui when using _Fairseq_ models. Argument `--speaker_wav` works fine on identical text with the XTTS model, but with Fairseq it seems to be ignored. Have tried both .wav and .mp3, different lengths, file locations/names, with and without CUDA, several languages. There are no errors, just always the same generic male voice. Is this a known issue with voice cloning and Fairseq on Windows’ command line or is something wrong with my setup?
### To Reproduce
_No response_
### Expected behavior
_No response_
### Logs
_No response_
### Environment
```shell
Windows, tts.exe
```
### Additional context
_No response_
|
closed
|
2023-11-05T17:02:20Z
|
2024-11-28T20:16:24Z
|
https://github.com/coqui-ai/TTS/issues/3142
|
[
"bug"
] |
Poccapx
| 21
|
ultralytics/ultralytics
|
computer-vision
| 19,550
|
Yolov11-12 tensorboard images section
|
### Search before asking
- [x] I have searched the Ultralytics YOLO [issues](https://github.com/ultralytics/ultralytics/issues) and [discussions](https://github.com/orgs/ultralytics/discussions) and found no similar questions.
### Question
Hello I'm collage student that using yolo models for object detection. I got stuck in somewhere. I have 10 labels for detection and these got pretty high mAP50 scores, but some labels are not as good scores as on the mAP50-95. For instance, I have traffic_light label. This label got 0.919 mAP50 score, but got 0.615 mAP50-95 score. Due to that, I want to see images that got false-true etc. on my data. I tried to use tensorboard but I couldn't view images section.
So here is my question:
**"Is there an any way to view the images with tensorboard ? "** if there isn possible, for this situation(higher mAP50, lower mAP50-95) what could I do ?. Thanks in advance.
### Additional
_No response_
|
open
|
2025-03-06T09:53:21Z
|
2025-03-11T17:57:57Z
|
https://github.com/ultralytics/ultralytics/issues/19550
|
[
"question",
"detect"
] |
MehmetKaTR
| 7
|
sanic-org/sanic
|
asyncio
| 2,982
|
Github Actions need updating
|
### Is there an existing issue for this?
- [X] I have searched the existing issues
### Is your feature request related to a problem? Please describe.
GitHub actions for publishing a package are on an old version, which uses a deprecated version of Node JS.
<img width="1109" alt="image" src="https://github.com/sanic-org/sanic/assets/25409753/757a829c-6a27-49cd-9cc4-ea9d40f6834b">
### Describe the solution you'd like
Update actions to latest versions.
### Additional context
_No response_
|
open
|
2024-06-30T12:38:42Z
|
2024-06-30T15:28:45Z
|
https://github.com/sanic-org/sanic/issues/2982
|
[] |
prryplatypus
| 3
|
davidteather/TikTok-Api
|
api
| 262
|
great job!! I want post a comment under a post,is there have any api for this?
|
**Is your feature request related to a problem? Please describe.**
A clear and concise description of what the problem is. Ex. I'm always frustrated when [...]
**Describe the solution you'd like**
A clear and concise description of what you want to happen.
**Describe alternatives you've considered**
A clear and concise description of any alternative solutions or features you've considered.
**Additional context**
Add any other context or screenshots about the feature request here.
|
closed
|
2020-09-14T15:44:09Z
|
2020-09-14T16:23:52Z
|
https://github.com/davidteather/TikTok-Api/issues/262
|
[
"feature_request"
] |
Gh-Levi
| 3
|
thunlp/OpenPrompt
|
nlp
| 140
|
Use 2.1_conditional_generation.py , after fine-tuning, it only generates the same char. Why ?
|
use 2.1_conditional_generation.py in datasets/CondGen/webnlg_2017/
generated txt: ''''''''''''''''''''''''''''''''''''''''''''''''''''''''''''''''''''''''''''''''''''''''''''''''''''''''''''''''
|
closed
|
2022-04-15T10:08:09Z
|
2022-05-30T09:14:56Z
|
https://github.com/thunlp/OpenPrompt/issues/140
|
[] |
353xiong
| 4
|
torchbox/wagtail-grapple
|
graphql
| 297
|
grapple vs wagtail-grapple
|
Initially I foundy myself confused by the difference between grapple and wagtail grapple. I had to pip install wagtail-grapple, but then import grapple in my code. It would be nice if the package only had one canonical name.
|
closed
|
2023-01-13T15:09:56Z
|
2024-09-20T09:49:55Z
|
https://github.com/torchbox/wagtail-grapple/issues/297
|
[] |
dopry
| 14
|
marcomusy/vedo
|
numpy
| 1,053
|
Is delete_cells_by_point_index parallelisable?
|
Current code:
```
def delete_cells_by_point_index(self, indices):
"""
Delete a list of vertices identified by any of their vertex index.
See also `delete_cells()`.
Examples:
- [delete_mesh_pts.py](https://github.com/marcomusy/vedo/tree/master/examples/basic/delete_mesh_pts.py)
"""
cell_ids = vtki.vtkIdList()
self.dataset.BuildLinks()
n = 0
for i in np.unique(indices):
self.dataset.GetPointCells(i, cell_ids)
for j in range(cell_ids.GetNumberOfIds()):
self.dataset.DeleteCell(cell_ids.GetId(j)) # flag cell
n += 1
self.dataset.RemoveDeletedCells()
self.dataset.Modified()
self.pipeline = OperationNode("delete_cells_by_point_index", parents=[self])
return self
```
Are there any issues with parallelising these two for loops? Even if it's via setting the number of jobs with joblib? It doesn't scale well (e.g. deleting half of a 150,000 point mesh). I'm not sure how VTK datasets work under the hood.
|
open
|
2024-02-16T02:52:30Z
|
2024-02-16T12:22:56Z
|
https://github.com/marcomusy/vedo/issues/1053
|
[] |
JeffreyWardman
| 1
|
matterport/Mask_RCNN
|
tensorflow
| 2,191
|
How to reduce inference detection time
|
I am using CPU for detection, when I run model.detect([image], verbose=1), it takes more than 25 seconds to detect for single image.
Is there any way to reduce the detection time?
|
open
|
2020-05-17T15:32:42Z
|
2020-11-19T10:31:00Z
|
https://github.com/matterport/Mask_RCNN/issues/2191
|
[] |
Dgs29
| 3
|
JaidedAI/EasyOCR
|
pytorch
| 856
|
TypeError: __init__() got an unexpected keyword argument 'detection'
|
Hi, I am trying to run this line ;
_reader = easyocr.Reader(['en'], detection='DB', recognition = 'Transformer')
But it is throwing me the following error;
TypeError: __init__() got an unexpected keyword argument 'detection'
|
closed
|
2022-09-17T19:06:30Z
|
2022-09-17T19:14:53Z
|
https://github.com/JaidedAI/EasyOCR/issues/856
|
[] |
sabaina-Haroon
| 1
|
deezer/spleeter
|
deep-learning
| 751
|
[Discussion] your question
|
ERROR: Cannot uninstall 'llvmlite'. It is a distutils installed project and thus we cannot accurately determine which files belong to it which would lead to only a partial uninstall.
|
closed
|
2022-04-17T21:48:13Z
|
2022-04-29T09:14:45Z
|
https://github.com/deezer/spleeter/issues/751
|
[
"question"
] |
sstefanovski21
| 0
|
LAION-AI/Open-Assistant
|
machine-learning
| 3,744
|
Can't open dashboard
| ERROR: type should be string, got "\r\nhttps://github.com/LAION-AI/Open-Assistant/assets/29770761/254db19d-a284-41ca-8612-99103df12fac\r\n\r\n"
|
closed
|
2024-01-06T16:14:36Z
|
2024-01-06T17:25:55Z
|
https://github.com/LAION-AI/Open-Assistant/issues/3744
|
[] |
DRYN07
| 1
|
benlubas/molten-nvim
|
jupyter
| 291
|
[Help] How to use cell magic like "%%time", "!ls" without pyright complaining?
|
First of all big thank you for this wonderful plugin. I'm trying to use cell magic like so:
<img width="726" alt="Image" src="https://github.com/user-attachments/assets/0c41a1ee-1e75-4429-be26-b8601487cac3" />
They would execute perfectly fine via ipython, but pyright complains about the syntax. Is there a way to suppress the error message?
On a side note that may be related, I'm not sure which plugin is causing this auto indent behavior after writing a cell magic:
https://github.com/user-attachments/assets/9db4d2b4-1339-4075-91ba-a8b3f58e2ae5
|
closed
|
2025-03-02T13:48:28Z
|
2025-03-04T17:12:45Z
|
https://github.com/benlubas/molten-nvim/issues/291
|
[] |
kanghengliu
| 6
|
kubeflow/katib
|
scikit-learn
| 2,149
|
Consolidate katib-cert-generator to katib-controller
|
/kind feature
**Describe the solution you'd like**
[A clear and concise description of what you want to happen.]
I would like to consolidate the katib-cert-generator to the katib-controller.
Currently, if we use the standalone cert-generator to generate self-signed certs for the webhooks, we can not use `Fail` as a failurePolicy for the `mutator.pod.katib.kubeflow.org` since we face the deadlock when we create the cert-generator pod via batch/job.
By generating the self-signed certs in katib-controller, we can avoid the above dead lock.
Ref: #2018
**Anything else you would like to add:**
[Miscellaneous information that will assist in solving the issue.]
---
<!-- Don't delete this message to encourage users to support your issue! -->
Love this feature? Give it a 👍 We prioritize the features with the most 👍
|
closed
|
2023-04-24T13:40:15Z
|
2023-08-04T19:31:23Z
|
https://github.com/kubeflow/katib/issues/2149
|
[
"kind/feature",
"release/0.16"
] |
tenzen-y
| 4
|
huggingface/diffusers
|
deep-learning
| 10,745
|
Unloading multiple loras: norms do not return to their original values
|
When unloading from multiple loras on flux pipeline, I believe that the norm layers are not restored [here](https://github.com/huggingface/diffusers/blob/464374fb87610c53b2cf81e08d3df628fada3ce4/src/diffusers/loaders/lora_pipeline.py#L1575).
Shouldn't we have:
```python
if len(transformer_norm_state_dict) > 0:
original_norm_layers_state_dict = self._load_norm_into_transformer(
transformer_norm_state_dict,
transformer=transformer,
discard_original_layers=False,
)
if not hasattr(transformer, "_transformer_norm_layers"):
transformer._transformer_norm_layers = original_norm_layers_state_dict
```
|
open
|
2025-02-07T15:43:12Z
|
2025-03-17T15:03:25Z
|
https://github.com/huggingface/diffusers/issues/10745
|
[
"stale"
] |
christopher5106
| 26
|
ultralytics/yolov5
|
pytorch
| 13,044
|
Parameters Fusion
|
### Search before asking
- [X] I have searched the YOLOv5 [issues](https://github.com/ultralytics/yolov5/issues) and [discussions](https://github.com/ultralytics/yolov5/discussions) and found no similar questions.
### Question
How to integrate some parameters from imported external modules into the entire YOLOv5 model for joint training?I want to introduce some filters as a module into the YOLOv5 model to enhance images. Input the original image of Yolov5 to the result of additional enhancement module, and the enhanced image is obtained in the first layer of the convolution block into Yolov5, and then trained together.How can I merge the parameters inside the filters into the trainable parameter list of YOLOv5 for joint training and updating?Thank you for help.
In common.py

In yaml

In train.py

### Additional
_No response_
|
closed
|
2024-05-28T08:24:22Z
|
2024-10-20T19:46:44Z
|
https://github.com/ultralytics/yolov5/issues/13044
|
[
"question",
"Stale"
] |
znmzdx-zrh
| 9
|
JaidedAI/EasyOCR
|
pytorch
| 1,028
|
RuntimeError: DataLoader worker (pid(s) 4308) exited unexpectedly
|
I get this error on training and don't know what is causing it
Traceback (most recent call last):
File "<string>", line 1, in <module>
File "C:\Users\FireAngelEmpire\anaconda3\lib\multiprocessing\spawn.py", line 116, in spawn_main
exitcode = _main(fd, parent_sentinel)
File "C:\Users\FireAngelEmpire\anaconda3\lib\multiprocessing\spawn.py", line 125, in _main
prepare(preparation_data)
File "C:\Users\FireAngelEmpire\anaconda3\lib\multiprocessing\spawn.py", line 236, in prepare
_fixup_main_from_path(data['init_main_from_path'])
File "C:\Users\FireAngelEmpire\anaconda3\lib\multiprocessing\spawn.py", line 287, in _fixup_main_from_path
main_content = runpy.run_path(main_path,
File "C:\Users\FireAngelEmpire\anaconda3\lib\runpy.py", line 289, in run_path
return _run_module_code(code, init_globals, run_name,
File "C:\Users\FireAngelEmpire\anaconda3\lib\runpy.py", line 96, in _run_module_code
_run_code(code, mod_globals, init_globals,
File "C:\Users\FireAngelEmpire\anaconda3\lib\runpy.py", line 86, in _run_code
exec(code, run_globals)
File "D:\situri\EasyOCRtrainer\trainer\main.py", line 37, in <module>
train(opt, amp=False)
File "D:\situri\EasyOCRtrainer\trainer\train.py", line 40, in train
train_dataset = Batch_Balanced_Dataset(opt)
File "D:\situri\EasyOCRtrainer\trainer\dataset.py", line 83, in __init__
self.dataloader_iter_list.append(iter(_data_loader))
File "C:\Users\FireAngelEmpire\anaconda3\lib\site-packages\torch\utils\data\dataloader.py", line 441, in __iter__
return self._get_iterator()
File "C:\Users\FireAngelEmpire\anaconda3\lib\site-packages\torch\utils\data\dataloader.py", line 388, in _get_iterator
return _MultiProcessingDataLoaderIter(self)
File "C:\Users\FireAngelEmpire\anaconda3\lib\site-packages\torch\utils\data\dataloader.py", line 1042, in __init__
w.start()
File "C:\Users\FireAngelEmpire\anaconda3\lib\multiprocessing\process.py", line 121, in start
self._popen = self._Popen(self)
File "C:\Users\FireAngelEmpire\anaconda3\lib\multiprocessing\context.py", line 224, in _Popen
return _default_context.get_context().Process._Popen(process_obj)
File "C:\Users\FireAngelEmpire\anaconda3\lib\multiprocessing\context.py", line 336, in _Popen
return Popen(process_obj)
File "C:\Users\FireAngelEmpire\anaconda3\lib\multiprocessing\popen_spawn_win32.py", line 45, in __init__
prep_data = spawn.get_preparation_data(process_obj._name)
File "C:\Users\FireAngelEmpire\anaconda3\lib\multiprocessing\spawn.py", line 154, in get_preparation_data
_check_not_importing_main()
File "C:\Users\FireAngelEmpire\anaconda3\lib\multiprocessing\spawn.py", line 134, in _check_not_importing_main
raise RuntimeError('''
RuntimeError:
An attempt has been made to start a new process before the
current process has finished its bootstrapping phase.
This probably means that you are not using fork to start your
child processes and you have forgotten to use the proper idiom
in the main module:
if __name__ == '__main__':
freeze_support()
...
The "freeze_support()" line can be omitted if the program
is not going to be frozen to produce an executable.
Traceback (most recent call last):
File "C:\Users\FireAngelEmpire\anaconda3\lib\site-packages\torch\utils\data\dataloader.py", line 1132, in _try_get_data
data = self._data_queue.get(timeout=timeout)
File "C:\Users\FireAngelEmpire\anaconda3\lib\queue.py", line 179, in get
raise Empty
_queue.Empty
The above exception was the direct cause of the following exception:
Traceback (most recent call last):
File "D:\situri\EasyOCRtrainer\trainer\main.py", line 37, in <module>
train(opt, amp=False)
File "D:\situri\EasyOCRtrainer\trainer\train.py", line 203, in train
image_tensors, labels = train_dataset.get_batch()
File "D:\situri\EasyOCRtrainer\trainer\dataset.py", line 101, in get_batch
image, text = next(data_loader_iter)
File "C:\Users\FireAngelEmpire\anaconda3\lib\site-packages\torch\utils\data\dataloader.py", line 633, in __next__
data = self._next_data()
File "C:\Users\FireAngelEmpire\anaconda3\lib\site-packages\torch\utils\data\dataloader.py", line 1328, in _next_data
idx, data = self._get_data()
File "C:\Users\FireAngelEmpire\anaconda3\lib\site-packages\torch\utils\data\dataloader.py", line 1284, in _get_data
success, data = self._try_get_data()
File "C:\Users\FireAngelEmpire\anaconda3\lib\site-packages\torch\utils\data\dataloader.py", line 1145, in _try_get_data
raise RuntimeError('DataLoader worker (pid(s) {}) exited unexpectedly'.format(pids_str)) from e
RuntimeError: DataLoader worker (pid(s) 4308) exited unexpectedly
I use default en_filtered_config.yaml settings
|
open
|
2023-05-23T22:58:06Z
|
2025-01-21T13:53:56Z
|
https://github.com/JaidedAI/EasyOCR/issues/1028
|
[] |
deadworldisee
| 1
|
kizniche/Mycodo
|
automation
| 1,313
|
Add "lock" feature to functions & outputs pages similar to Dashboard "lock" button function.
|
Sometimes when accessing the Mycodo web GUI from a phone, functions and outputs can accidentally get moved and jumbled if the drag handles are touched when trying to scroll the screen.
Would it be possible to add a lock feature like on the Dashboard pages that prevents any of the widgets from being moved?
I now have a Mycodo system running that is being accessed by multiple users, and it would be nice to lock the Function and Output screens the same way I can lock the Dashboards to prevent them from getting jumbled by accident... or from changing layout when viewed on a different resolution browser.
|
open
|
2023-06-13T08:09:19Z
|
2023-08-11T03:40:47Z
|
https://github.com/kizniche/Mycodo/issues/1313
|
[
"enhancement"
] |
LucidEye
| 1
|
ultralytics/ultralytics
|
machine-learning
| 19,838
|
where is yolo3D?
|
### Search before asking
- [x] I have searched the Ultralytics [issues](https://github.com/ultralytics/ultralytics/issues) and found no similar feature requests.
### Description
where is yolo3D?
### Use case
_No response_
### Additional
_No response_
### Are you willing to submit a PR?
- [ ] Yes I'd like to help by submitting a PR!
|
open
|
2025-03-24T08:35:01Z
|
2025-03-24T10:43:16Z
|
https://github.com/ultralytics/ultralytics/issues/19838
|
[
"enhancement",
"question"
] |
xinsuinizhuan
| 2
|
CPJKU/madmom
|
numpy
| 92
|
refactor add_arguments of all FilteredSpectrogramProcessor and MultiBandSpectrogramProcessor
|
Most of the duplicated code could be refactored to `audio.filters`.
|
closed
|
2016-02-18T12:56:16Z
|
2016-02-24T08:21:30Z
|
https://github.com/CPJKU/madmom/issues/92
|
[] |
superbock
| 0
|
iterative/dvc
|
data-science
| 9,891
|
dvc data status: handle broken symlinks to cache
|
Managing shared caches is difficult, and so sometimes we are naughty and just delete data from the cache directly. (This is much easier than trying to manage `dvc gc -p`.)
The result is dangling symlinks, and `dvc data status` trips over these with ...
```
ERROR: unexpected error - [Errno 2] No such file or directory
```
It would be nice to either have an `--allow-missing` option, so that `dvc data status` processes what _is_ there, or perhaps `dvc data status` could handle these situations as "not in cache", which would be a fair description.
|
open
|
2023-08-31T04:51:52Z
|
2023-09-06T03:26:34Z
|
https://github.com/iterative/dvc/issues/9891
|
[] |
johnyaku
| 2
|
nvbn/thefuck
|
python
| 1,177
|
Feature Request: rvm
|
Would be great to add support for rvm, such as in the following example:
```
mensly ~> rvm use 2.7.2
Required ruby-2.7.2 is not installed.
To install do: 'rvm install "ruby-2.7.2"'
mensly ~> fuck
No fucks given
mensly ~> rvm install "ruby-2.7.2"
```
|
open
|
2021-03-22T02:41:38Z
|
2021-07-23T16:31:17Z
|
https://github.com/nvbn/thefuck/issues/1177
|
[
"help wanted",
"hacktoberfest"
] |
mensly
| 0
|
jupyter-book/jupyter-book
|
jupyter
| 1,912
|
Footnote does not show up with Utterances.
|
### Describe the bug
Hi, I wish this is not a duplicated issue and I am sorry for my poor English in advance.
**context**
I have used Utterances as commenting service on my Jupyter Book and I just found out that a footnote does not show up with Utterances.
I would like to note that I have manually inserted Utterances into each page by using the following code (which can be found [here](https://github.com/executablebooks/jupyter-book/blob/master/docs/interactive/comments/utterances.md#configure-utterances)) because Utterances had not showed up when I followed the [instruction](https://jupyterbook.org/en/stable/interactive/comments/utterances.html#utterances).
~~~
```{raw} html
<script
type="text/javascript"
src="https://utteranc.es/client.js"
async="async"
repo="HiddenBeginner/Deep-Reinforcement-Learnings"
issue-term="pathname"
theme="github-light"
label="💬 comment"
crossorigin="anonymous"
/>
```
~~~
### Reproduce the bug
The followings are one of my pages that contain footnotes and the corresponding Markdown/HTML files. I apologize this is not an English page. As you can see, I tried to add a footnote at the word "SOTA", but it did not show up. When I removed codes for embedding Utterance, the footnote showed up properly.
- **Page**: https://hiddenbeginner.github.io/Deep-Reinforcement-Learnings/book/intro.html
- **Markdown**: https://github.com/HiddenBeginner/Deep-Reinforcement-Learnings/blob/master/book/intro.md
- **HTML**: https://github.com/HiddenBeginner/Deep-Reinforcement-Learnings/blob/gh-pages/book/intro.html#L396
- **_config.yml**: https://github.com/HiddenBeginner/Deep-Reinforcement-Learnings/blob/master/_config.yml
### List your environment
```
> jupyter-book --version
Jupyter Book : 0.13.1
External ToC : 0.2.4
MyST-Parser : 0.15.2
MyST-NB : 0.13.2
Sphinx Book Theme : 0.3.3
Jupyter-Cache : 0.4.3
NbClient : 0.5.13
```
Thank you in advance :).
|
open
|
2023-01-21T04:22:04Z
|
2023-01-24T06:45:16Z
|
https://github.com/jupyter-book/jupyter-book/issues/1912
|
[
"bug"
] |
HiddenBeginner
| 1
|
slackapi/bolt-python
|
fastapi
| 350
|
Problem with globals in Socket Mode + Flask?
|
Hi! I use SocketMode in my application in conjunction with Flask, in order to make a health check of the probes, so I use in this form:
```bash
@flask_app.route('/readiness')
def readiness():
return {"status": "Ok"}
if __name__ == "__main__":
SocketModeHandler(app, settings.slack_app_token).connect()
flask_app.run(host="0.0.0.0", port=14111, threaded=True, debug=True)
```
but in this case, for some reason, my global variables in the application are periodically lost between calls to actions.
When I use it like this without Flask, everything works fine, but I need this health check probe
```bash
if __name__ == "__main__":
SocketModeHandler(app, settings.slack_app_token).start()
```
What could be the reason? And how can you get around this?
#### The `slack_bolt` version
slack-bolt==1.4.4
slack-sdk==3.4.2
#### Python runtime version
Python 3.8.2
#### OS info
ProductName: Mac OS X
ProductVersion: 10.15.7
BuildVersion: 19H2
Darwin Kernel Version 19.6.0
|
closed
|
2021-05-25T13:34:29Z
|
2021-06-19T01:52:34Z
|
https://github.com/slackapi/bolt-python/issues/350
|
[
"question",
"area:adapter"
] |
De1f364
| 7
|
tflearn/tflearn
|
tensorflow
| 987
|
Interface for 3D max pooling inconsistent.
|
`tflearn.layers.conv.max_pool_1d` and `tflearn.layers.conv.max_pool_2d` default strides is equal to the kernel size. For`tflearn.layers.conv.max_pool_3d` this is not the case, there the default strides equals to 1.
While not really a bug this could lead to confusion with people which are used to the 1d and 2d interfaces and do not realize that the interface is different in the 3d version.
|
open
|
2017-12-30T09:48:31Z
|
2017-12-30T09:49:14Z
|
https://github.com/tflearn/tflearn/issues/987
|
[] |
deeplearningrobotics
| 0
|
jowilf/starlette-admin
|
sqlalchemy
| 391
|
Enhancement: Customizable profile menu
|
**Is your feature request related to a problem? Please describe.**
The profile menu has only one button: logout. I'm proposing to implement a way to extend this menu from the python side. Maybe a hook like `auth_provider`. `profile_menu` could have a bunch of items in it and we could define the functionality with `expose` feature that I proposed in #389
**Additional context**
I'm planning to do some work on this, wdyt about this feature @jowilf?
|
closed
|
2023-11-08T09:19:11Z
|
2023-12-04T00:53:33Z
|
https://github.com/jowilf/starlette-admin/issues/391
|
[
"enhancement"
] |
hasansezertasan
| 0
|
Evil0ctal/Douyin_TikTok_Download_API
|
fastapi
| 452
|
[BUG] 一个星期前使用Web app过并且成功下载了200+视频,但是这个星期突然不行了,按照视频教程更换了Cookie,仍然不行
|
***发生错误的平台?***
如:抖音
***发生错误的端点?***
如:Web APP
***提交的输入值?***
如:短视频链接
***是否有再次尝试?***
如:是,发生错误后X时间后错误依旧存在。
***你有查看本项目的自述文件或接口文档吗?***
如:有,并且很确定该问题是程序导致的。
这是我的Cookie,已经按照了视频教程重复过多次,并且在一个星期前已经成功下载了200+视频,但是不知道为什么最近这几天突然出现视频教程中的情况突然不能解析了,有可能是我的Cookie除了问题吗?
ttwid=1%7C-0dflMMeBDApYTLvDCHU5M9zmhMbAw9bQwL1B888aeQ%7C1721186940%7C5f2afca82abcb491efe8c7e89507eb2efd7065db436ad029b9968907e3204dfe; UIFID_TEMP=3c3e9d4a635845249e00419877a3730e2149197a63ddb1d8525033ea2b3354c2b36d8dea70ec55ef3bfed51712fd79effc967048ca658629071e58a611b087b705e805e59215e9bd4f3f99e55df3d64d; strategyABtestKey=%221721186954.034%22; FORCE_LOGIN=%7B%22videoConsumedRemainSeconds%22%3A180%7D; passport_csrf_token=74f7f4e43ed3f216c16e4796982cdf05; passport_csrf_token_default=74f7f4e43ed3f216c16e4796982cdf05; bd_ticket_guard_client_web_domain=2; UIFID=3c3e9d4a635845249e00419877a3730e2149197a63ddb1d8525033ea2b3354c2262f6b2f53dc3e825dd4d5b94b2c1b7e271861bbbf8ec76081e02ae101abda49170744221a288426c30f82755020708ebdb1479ed9405f571b2e832c09476249b27dc14a5e112b9987dfedeb2710df8fd409b287156739d0644f0bea9f02e0376d32a2eaf2a7630384bf1373393aeb43f580dae88956ae8b798841594723fce1; publish_badge_show_info=%220%2C0%2C0%2C1721187009386%22; _bd_ticket_crypt_doamin=2; __security_server_data_status=1; store-region-src=uid; volume_info=%7B%22isUserMute%22%3Afalse%2C%22isMute%22%3Atrue%2C%22volume%22%3A0.5%7D; stream_player_status_params=%22%7B%5C%22is_auto_play%5C%22%3A0%2C%5C%22is_full_screen%5C%22%3A0%2C%5C%22is_full_webscreen%5C%22%3A0%2C%5C%22is_mute%5C%22%3A1%2C%5C%22is_speed%5C%22%3A1%2C%5C%22is_visible%5C%22%3A0%7D%22; download_guide=%223%2F20240717%2F0%22; WallpaperGuide=%7B%22showTime%22%3A1721197085102%2C%22closeTime%22%3A0%2C%22showCount%22%3A1%2C%22cursor1%22%3A12%2C%22cursor2%22%3A0%7D; pwa2=%220%7C0%7C3%7C0%22; stream_recommend_feed_params=%22%7B%5C%22cookie_enabled%5C%22%3Atrue%2C%5C%22screen_width%5C%22%3A1920%2C%5C%22screen_height%5C%22%3A1080%2C%5C%22browser_online%5C%22%3Atrue%2C%5C%22cpu_core_num%5C%22%3A6%2C%5C%22device_memory%5C%22%3A8%2C%5C%22downlink%5C%22%3A1.3%2C%5C%22effective_type%5C%22%3A%5C%223g%5C%22%2C%5C%22round_trip_time%5C%22%3A400%7D%22; d_ticket=d3be1d1c514794e148e688b89e54f96380010; passport_assist_user=CkDm--hTvOv3nEIRYcioVyfALzX8qEAQEtREu2tsz6MopT5gQZjQb61iJsUpB4XmasV7N1vOMO0wf7eXqpkBBHkOGkoKPHfDqDGzaj8jgaZ3v9CbokP7pgRqx5kPzx0j_ruHplQ5i0w-UcKgL2arE5Q-yPDkskxtqJcCPOn7qwL-yRC38tYNGImv1lQgASIBAxZNPMs%3D; n_mh=CXji1rShkMmP9Poc3bNuzzGe8F_66lf_NsDkrYBa_Ok; sso_uid_tt=ff2232aa2246a526d1f5688fc341e51c; sso_uid_tt_ss=ff2232aa2246a526d1f5688fc341e51c; toutiao_sso_user=6bac0b1120e173cf26cf7982ef052dd0; toutiao_sso_user_ss=6bac0b1120e173cf26cf7982ef052dd0; sid_ucp_sso_v1=1.0.0-KDc0ODA3ZWZjMmFmYzFlMGU1NGE2NTk5Mjk2Njc3NDVjNTUyZjc0NzIKIAiP8uDD5Y0qELj73bQGGO8xIAww3I-WjwY4BkD0B0gGGgJobCIgNmJhYzBiMTEyMGUxNzNjZjI2Y2Y3OTgyZWYwNTJkZDA; ssid_ucp_sso_v1=1.0.0-KDc0ODA3ZWZjMmFmYzFlMGU1NGE2NTk5Mjk2Njc3NDVjNTUyZjc0NzIKIAiP8uDD5Y0qELj73bQGGO8xIAww3I-WjwY4BkD0B0gGGgJobCIgNmJhYzBiMTEyMGUxNzNjZjI2Y2Y3OTgyZWYwNTJkZDA; passport_auth_status=64b41651d22ef01c3bd7caabc5b4a3c3%2Cbd35a4f84b65debf4e03b65971e4218b; passport_auth_status_ss=64b41651d22ef01c3bd7caabc5b4a3c3%2Cbd35a4f84b65debf4e03b65971e4218b; uid_tt=fc9cc028b2baed1909e2390fb7b91756; uid_tt_ss=fc9cc028b2baed1909e2390fb7b91756; sid_tt=6e1976c2fda4e56d99ac2ed7092ef80e; sessionid=6e1976c2fda4e56d99ac2ed7092ef80e; sessionid_ss=6e1976c2fda4e56d99ac2ed7092ef80e; IsDouyinActive=true; FOLLOW_NUMBER_YELLOW_POINT_INFO=%22MS4wLjABAAAAXp9b19ZnYlvq_ENlNiL4OXsFHV2k2je9Yt1CbGCjJf0%2F1721232000000%2F0%2F1721204159595%2F0%22; home_can_add_dy_2_desktop=%221%22; _bd_ticket_crypt_cookie=c1acf69845fdeffea2b61ab4b4860e20; bd_ticket_guard_client_data=eyJiZC10aWNrZXQtZ3VhcmQtdmVyc2lvbiI6MiwiYmQtdGlja2V0LWd1YXJkLWl0ZXJhdGlvbi12ZXJzaW9uIjoxLCJiZC10aWNrZXQtZ3VhcmQtcmVlLXB1YmxpYy1rZXkiOiJCSGN0Z29QdlYrMER4MmR5TGVXcTl4cmY2UHlCR2syeENMZDVBNXhJakVxblBRa2Fham44dkhxL0NTOHpkR3lZNzIwUGp2YW90UkpnR3BRdW11K1RkVGM9IiwiYmQtdGlja2V0LWd1YXJkLXdlYi12ZXJzaW9uIjoxfQ%3D%3D; sid_guard=6e1976c2fda4e56d99ac2ed7092ef80e%7C1721204164%7C5183991%7CSun%2C+15-Sep-2024+08%3A15%3A55+GMT; sid_ucp_v1=1.0.0-KGQ4MjQ1YTAxYjE3M2Y3MDJhMzc5N2U3MmE5ZjJkMDFmY2IxZWJiNTIKGgiP8uDD5Y0qEMT73bQGGO8xIAw4BkD0B0gEGgJobCIgNmUxOTc2YzJmZGE0ZTU2ZDk5YWMyZWQ3MDkyZWY4MGU; ssid_ucp_v1=1.0.0-KGQ4MjQ1YTAxYjE3M2Y3MDJhMzc5N2U3MmE5ZjJkMDFmY2IxZWJiNTIKGgiP8uDD5Y0qEMT73bQGGO8xIAw4BkD0B0gEGgJobCIgNmUxOTc2YzJmZGE0ZTU2ZDk5YWMyZWQ3MDkyZWY4MGU; biz_trace_id=7e842496; store-region=cn-gd; odin_tt=be8a61af146d51affbe8ab19ff575abc11b795bef9755e544d0cc8410ac6ee759197ac56b90a0e262a033ebc23c7114a29c14ae6010dd3459af6a82a58ba659f
|
closed
|
2024-07-17T08:47:35Z
|
2024-07-27T21:53:09Z
|
https://github.com/Evil0ctal/Douyin_TikTok_Download_API/issues/452
|
[
"BUG"
] |
Losoy1
| 6
|
deeppavlov/DeepPavlov
|
tensorflow
| 876
|
DeepPavlov Bert 10x slower than Pytorch Pretrained Bert
|
Hi,
I modified the default DeepPavlov Bert to only output the pooled output (I followed the instructions given in [my previous issue](https://github.com/deepmipt/DeepPavlov/issues/825)).
However, this modified DeepPavlov Bert version is 10x slower than the [Pytorch Pretrained Bert](https://github.com/huggingface/pytorch-pretrained-BERT).
The code used for DeepPavlov Bert:
```
text = "Once when I was six years old I saw a magnificent picture in a book, called True Stories from Nature, about the primeval forest. It was a picture of a boa constrictor in the act of swallowing an animal. Here is a copy of the drawing."
model = deeppavlov.build_model('tools/deeppavlov/squad_bert.json')
embedding = model([text], [''])[0]
```
It takes 0.85 second to get the embedding.
Moreover, `model([text*10], [''])[0]` only takes 1 second, whereas `model([text]*10, ['']*10)[0]` takes 4 seconds. This should take the same time, isn't it?
|
closed
|
2019-06-11T13:09:42Z
|
2020-05-13T09:48:54Z
|
https://github.com/deeppavlov/DeepPavlov/issues/876
|
[] |
lcswillems
| 1
|
pydata/pandas-datareader
|
pandas
| 453
|
RLS 0.6.0
|
There have been a lot of changes in the past few days.
Please report any issues here. I there are none raised by the start of next week, 0.6.0 will be released then.
- [x] Add release date to what's new
- [x] Tag release
|
closed
|
2018-01-18T16:36:28Z
|
2018-01-29T21:42:08Z
|
https://github.com/pydata/pandas-datareader/issues/453
|
[] |
bashtage
| 24
|
opengeos/streamlit-geospatial
|
streamlit
| 39
|
issue
|
how to download data from this app
and how to add districts,towns,etc
how to add lat and long from live APi
|
closed
|
2022-04-12T16:39:32Z
|
2022-05-19T13:15:40Z
|
https://github.com/opengeos/streamlit-geospatial/issues/39
|
[] |
Zar-Jamil
| 1
|
lundberg/respx
|
pytest
| 138
|
Consider changing the badges
|
[](https://github.com/lundberg/respx/actions/workflows/test.yml) [](https://codecov.io/gh/lundberg/respx) [](https://pypi.org/project/respx/) [](https://pypi.org/project/respx/)
|
closed
|
2021-03-03T21:13:58Z
|
2021-07-06T08:37:35Z
|
https://github.com/lundberg/respx/issues/138
|
[] |
lundberg
| 0
|
ets-labs/python-dependency-injector
|
asyncio
| 865
|
Implement Python dependency injector library in Azure Functions
|
### Description of Issue
I am trying to implement the dependency injector for Python Azure functions.
i tried to implement it using a Python library called Dependency Injector.
pip install dependency-injector [https://python-dependency-injector.ets-labs.org](https://python-dependency-injector.ets-labs.org/)
However, I am getting below error.
> Error: "functions.http_app_func. System.Private.CoreLib: Result: Failure Exception: AttributeError: 'dict' object has no attribute 'encode'"
This is the code I am trying to implement. Please have someone guide me here.
> function app file name: function_app.py
```
import azure.functions as func
from fastapi import FastAPI, Depends, Request, Response
from dependency_injector.wiring import inject, Provide
from abstraction.di_container import DIContainer
import logging
import json
from src.config.app_settings import AppSettings
container = DIContainer()
container.wire(modules=[__name__])
fast_app = FastAPI()
@fast_app.exception_handler(Exception)
async def handle_exception(request: Request, exc: Exception):
return Response(
status_code=400,
content={"message": str(exc)},
)
@fast_app.get("/")
@inject
async def home(settings: AppSettings = Depends(Provide[DIContainer.app_config])):
cont_name = settings.get("ContainerName", "No setting found")
return {
"info": f"Try to get values from local.settings using DI {cont_name}"
}
@fast_app.get("/v1/test/{test}")
async def get_test(self,
test: str):
return {
"test": test
}
app = func.AsgiFunctionApp(app=fast_app, http_auth_level=func.AuthLevel.ANONYMOUS)
```
> Dependency Injector file name: di_container.py
```
from dependency_injector import containers, providers
from src.config.app_settings import AppSettings
class DIContainer(containers.DeclarativeContainer):
app_config = providers.Singleton(AppSettings)
```
> Application Setting to read local.settings.json file: app_settings.py
```
import json
import os
from dependency_injector import containers, providers
class AppSettings:
def __init__(self, file_path="local.settings.json"):
self.config_data = {}
if os.path.exists(file_path):
with open(file_path, "r", encoding="utf-8") as file:
data = json.load(file)
self.config_data = data.get("Values", {})
def get(self, key: str, default=None):
return os.getenv(key,self.config_data.get(key, default))
```
|
open
|
2025-03-03T05:29:25Z
|
2025-03-03T05:29:25Z
|
https://github.com/ets-labs/python-dependency-injector/issues/865
|
[] |
Ramkisubramanian
| 0
|
pydantic/logfire
|
fastapi
| 813
|
Cannot incorporate VCS root_path
|
I am struggling to get the root_path component of my VCS configuration to work. I've tried configuring via `logfire.CodeSource()` as well as setting the `OTEL_RESOURCE_ATTRIBUTES` environment variable (not at the same time).
My current configuration is as follows -
```
logfire.configure(
environment=os.environ["ENVIRONMENT"],
code_source=logfire.CodeSource(
repository=os.environ["LOGFIRE_REPOSITORY"],
revision=os.environ["LOGFIRE_REVISION"],
root_path=os.environ["LOGFIRE_ROOT_PATH"],
),
)
```
The root directory of my code is in a subdirectory mounted in my Docker container at - /repository_root_path/code_subdirectory
And my actual python code runs out of - /repository_root_path/code_subdirectory/src
I've set the variables as the following -
```
LOGFIRE_REPOSITORY=https://github.com/my-org/my-repo
LOGFIRE_REVISION=main
LOGFIRE_ROOT_PATH=code_subdirectory
```
The resulting links generated in Logfire follow this format - https://github.com/my-org/my-repo/blob/main/src/path/to/code
Whereas the valid link is - https://github.com/my-org/my-repo/blob/main/code_subdirectory/src/path/to/code
Upon further review, it seems like changing the `root_path` doesn't impact the links at all so I'm not sure what to do.
|
open
|
2025-01-21T16:14:51Z
|
2025-01-23T16:28:00Z
|
https://github.com/pydantic/logfire/issues/813
|
[] |
fwinokur
| 6
|
PaddlePaddle/PaddleHub
|
nlp
| 2,195
|
paddlehub 引用sklearn 报 cannot load any more object with static TLS 错误
|
系统环境
paddle.__version__ 2.4.0-rc0
#### 具体代码
!/usr/bin/env python
#-*- coding=utf8 -*-
import os
import sys
import paddlehub as hub
module = hub.Module(name="lac")
test_text = '小明硕士毕业于中国科学院计算所,后在日本京都大学深造'
results = module.lexical_analysis(texts=test_text)
###############
Traceback (most recent call last):
File "/ssd4/liuyaping/python38/lib/python3.8/site-packages/sklearn/__check_build/__init__.py", line 44, in <module>
from ._check_build import check_build # noqa
ImportError: dlopen: cannot load any more object with static TLS
During handling of the above exception, another exception occurred:
Traceback (most recent call last):
File "test_lac.py", line 5, in <module>
import paddlehub as hub
File "/ssd4/liuyaping/python38/lib/python3.8/site-packages/paddlehub/__init__.py", line 31, in <module>
from paddlehub import datasets
File "/ssd4/liuyaping/python38/lib/python3.8/site-packages/paddlehub/datasets/__init__.py", line 16, in <module>
from paddlehub.datasets.chnsenticorp import ChnSentiCorp
File "/ssd4/liuyaping/python38/lib/python3.8/site-packages/paddlehub/datasets/chnsenticorp.py", line 19, in <module>
from paddlehub.datasets.base_nlp_dataset import TextClassificationDataset
File "/ssd4/liuyaping/python38/lib/python3.8/site-packages/paddlehub/datasets/base_nlp_dataset.py", line 21, in <module>
import paddlenlp
File "/ssd4/liuyaping/python38/lib/python3.8/site-packages/paddlenlp/__init__.py", line 29, in <module>
from . import metrics
File "/ssd4/liuyaping/python38/lib/python3.8/site-packages/paddlenlp/metrics/__init__.py", line 16, in <module>
from .chunk import ChunkEvaluator
File "/ssd4/liuyaping/python38/lib/python3.8/site-packages/paddlenlp/metrics/chunk.py", line 6, in <module>
from seqeval.metrics.sequence_labeling import get_entities
File "/ssd4/liuyaping/python38/lib/python3.8/site-packages/seqeval/metrics/__init__.py", line 1, in <module>
from seqeval.metrics.sequence_labeling import (accuracy_score,
File "/ssd4/liuyaping/python38/lib/python3.8/site-packages/seqeval/metrics/sequence_labeling.py", line 14, in <module>
from seqeval.metrics.v1 import SCORES, _precision_recall_fscore_support
File "/ssd4/liuyaping/python38/lib/python3.8/site-packages/seqeval/metrics/v1.py", line 5, in <module>
from sklearn.exceptions import UndefinedMetricWarning
File "/ssd4/liuyaping/python38/lib/python3.8/site-packages/sklearn/__init__.py", line 79, in <module>
from . import __check_build # noqa: F401
File "/ssd4/liuyaping/python38/lib/python3.8/site-packages/sklearn/__check_build/__init__.py", line 46, in <module>
raise_build_error(e)
File "/ssd4/liuyaping/python38/lib/python3.8/site-packages/sklearn/__check_build/__init__.py", line 31, in raise_build_error
raise ImportError("""%s
ImportError: dlopen: cannot load any more object with static TLS
___________________________________________________________________________
Contents of /ssd4/liuyaping/python38/lib/python3.8/site-packages/sklearn/__check_build:
_check_build.cpython-38-x86_64-linux-gnu.sosetup.py __pycache__
__init__.py
___________________________________________________________________________
It seems that scikit-learn has not been built correctly.
|
open
|
2023-01-12T06:42:38Z
|
2023-01-13T09:38:14Z
|
https://github.com/PaddlePaddle/PaddleHub/issues/2195
|
[] |
hnlslyp
| 1
|
JaidedAI/EasyOCR
|
machine-learning
| 746
|
Export to ONNX and use ONNX Runtime, working. Guide.
|
This is an explanation of how to export the recognition model and the detection model to ONNX format. Then a brief explanation of how to use ONNX Runtime to use these models.
ONNX is an intercompatibility standard for AI models. It allows us to use the same model in different types of programming languages, operating systems, acceleration platforms and runtimes. Personally I need to make a C++ build of EasyOCR functionality. After failing, due to several reasons, to make a C++ build using Pytorch and the EasyOCR models, I found that the best solution is to transform the models to ONNX and then program in C++ using ONNX Runtime. Then, compiling is very easy compared to PyTorch.
Due to time constraints I am not presenting a PR. It will be necessary for you to modify a copy of EasyOCR locally.
## Requirements
We must install the modules: [onnx](https://github.com/onnx/onnx) and [onnxruntime](https://onnxruntime.ai/docs/get-started/with-python.html). In my case I also had to manually install the [protobuf](https://pypi.org/project/protobuf/) module in version **3.20**.
I am using:
- EasyOCR 1.5.0
- Python 3.9.9
- torch 1.10.1
- torchvision 0.11.2
- onnx 1.11.0
- onnxruntime 1.11.1
## Exporting ONNX models
The best place to modify the EasyOCR code to export the models is right after EasyOCR uses the loaded model to perform the prediction.
### Exporting detection model
In `easyocr/detection.py` after `y, feature = net(x)` (line 46) add:
```
batch_size_1 = 500
batch_size_2 = 500
in_shape=[1, 3, batch_size_1, batch_size_2]
dummy_input = torch.rand(in_shape)
dummy_input = dummy_input.to(device)
torch.onnx.export(
net.module,
dummy_input,
"detectionModel.onnx",
export_params=True,
opset_version=11,
input_names = ['input'],
output_names = ['output'],
dynamic_axes={'input' : {2 : 'batch_size_1', 3: 'batch_size_2'}},
)
```
We generate a dumb input, totally random, so that onnx can perform the export. It doesn't matter the input, the important thing is that it has the correct structure. The detection model uses an input that is a 4-dimensional tensor, where the first dimension always has a value of 1, the second a value of 3 and the third and fourth values depend on the resolution of the analyzed image. I have assumed this conclusion after analyzing the data flow, I may be in error and this needs to be corrected.
Note that we export with the parameters (`export_params=True`) and specify that the two final dimensions of the input tensor are of dynamic size (`dynamic_axes=...`).
Then we can add this code to immediately import the exported model and validate that it is not corrupted:
```
onnx_model = onnx.load("detectionModel.onnx")
try:
onnx.checker.check_model(onnx_model)
except onnx.checker.ValidationError as e:
print('The model is invalid: %s' % e)
else:
print('The model is valid!')
```
Remember to `import onnx` in the file header.
To run the export just use EasyOCR and perform an analysis on any image indicating the language to be detected. This will download the corresponding model, run the detection and simultaneously export the model. If we change the language we will have to export a new model. Once the model is exported, we can comment or delete the code.
### Exporting the recognition model
This model is a bit more difficult to export and we will have to do some black magic.
In `easyocr/recognition.py` after `preds = model(image, text_for_pred)` (line 111) add:
```
batch_size_1_1 = 500
in_shape_1=[1, 1, 64, batch_size_1_1]
dummy_input_1 = torch.rand(in_shape_1)
dummy_input_1 = dummy_input_1.to(device)
batch_size_2_1 = 50
in_shape_2=[1, batch_size_2_1]
dummy_input_2 = torch.rand(in_shape_2)
dummy_input_2 = dummy_input_2.to(device)
dummy_input = (dummy_input_1, dummy_input_2)
torch.onnx.export(
model.module,
dummy_input,
"recognitionModel.onnx",
export_params=True,
opset_version=11,
input_names = ['input1','input2'],
output_names = ['output'],
dynamic_axes={'input1' : {3 : 'batch_size_1_1'}},
)
```
As with the detection model, we create a dumb input to be able to export the model. In this case, the model input is 2 elements.
The first element is a 4-dimensional tensor, where the first dimension always has a value of 1, the second a value of 1, the third a value of 64 and the fourth a dynamic value.
The second element is a 2-dimensional tensor, where the first dimension always has a value of 1 and the second a dynamic value.
Again, I may be wrong about the structure of these inputs, it was what I observed empirically.
**First strange thing:** ONNX for some reason, in performing its analysis of the model structure, concludes that the second input element does not perform any function. So even if we tell ONNX to export a model with 2 input elements, it will always export a model with 1 input element. It appears that this is due to an internal ONNX process where it "cuts" parts of the network defining graph that do not alter the network output. According to the documentation we can stop this "cutting" process and export the network without optimization using the `do_constant_folding=False` parameter as an option. [But due to a bug](https://github.com/pytorch/pytorch/issues/44299) it is not taking effect. In spite of the above, we can observe that this lack of the second element does not generate losses in the accuracy of the model. For this reason, in the dynamic elements (`dynamic_axes=`) we only define one element where its third dimension is variable in size. If anyone manages to export the model with the two input elements, it would be appreciated if you could notify us.
**Second strange thing:** In order to export the recognition model, we must edit `easyocr/model/vgg_model.py`. It turns out that the [AdaptiveAvgPool2d](https://pytorch.org/docs/stable/generated/torch.nn.AdaptiveAvgPool2d.html) operator is not fully [supported](https://github.com/onnx/onnx/blob/main/docs/Operators.md) by ONNX. **When it uses the "None" option**, in the configuration tuple (which indicates that the size must be equal to the input), the export fails. To fix this we need to change line 11:
From
`self.AdaptiveAvgPool = nn.AdaptiveAvgPool2d((None, 1))`
to
`self.AdaptiveAvgPool = nn.AdaptiveAvgPool2d((256, 1))`
Why 256? I don't know. Is there a better option? I have not found one. Does it generate errors in the model? I have not been able to find any accuracy problems. If someone can explain why with 256 it works and what the consequences are, it would be appreciated.
Well then, just like the detection model we can add these lines to validate the exported model:
```
onnx_model = onnx.load("detectionModel.onnx")
try:
onnx.checker.check_model(onnx_model)
except onnx.checker.ValidationError as e:
print('The model is invalid: %s' % e)
else:
print('The model is valid!')
```
Remember to `import onnx` in the file header.
To export the recognition model we must run EasyOCR using any image and the desired language. In the process you will see that some alerts will be generated, but you can ignore them. The model will be exported several times, since the added code has been placed inside a for loop. But this should not cause any problems. Remember to comment or remove the added code afterwards. If you change language, you must export a new ONNX model.
## Using ONNX models in EasyOCR
To test and validate that the models work, we will modify the code again. This time we will comment the lines where EasyOCR uses the Pytorch prediction and we will add the code to use ONNX Runtime to perform the prediction.
### Using the ONNX detection model
First we must add this helper function to the file `easyocr/detection.py`:
```
def to_numpy(tensor):
return tensor.detach().cpu().numpy() if tensor.requires_grad else tensor.cpu().numpy()
```
Then we must comment on linear 46 where it says `y, feature = net(x)`. After this line we must add:
```
ort_session = onnxruntime.InferenceSession("detectionModel.onnx")
ort_inputs = {ort_session.get_inputs()[0].name: to_numpy(x)}
ort_outs = ort_session.run(None, ort_inputs)
y = ort_outs[0]
```
Remember to `import onnxruntime` in the file header.
In this way we load the ONNX model of detection and pass as input the value "x". Since ONNX does not use Pytorch, we must convert "x" from a Tensor to a standard numpy array. Para eso usamos la función de ayuda The output of ONNX is left in the "y" variable.
One last modification must be made on lines 51 and 52. Change from:
```
score_text = out[:, :, 0].cpu().data.numpy()
score_link = out[:, :, 1].cpu().data.numpy()
```
to
```
score_text = out[:, :, 0]
score_link = out[:, :, 1]
```
This is because the model output is already a numpy array and does not need to be converted from a Tensor.
**To test, we can run EasyOCR with some image and see the result.**
### Using the ONNX recognition model
We must add the help function to the file `easyocr/recognition.py`:
```
def to_numpy(tensor):
return tensor.detach().cpu().numpy() if tensor.requires_grad else tensor.cpu().numpy()
```
Then we must comment on linear 111 to stop using PyTorch prediction: `preds = model(image, text_for_pred)`. And right after that add:
```
ort_session = onnxruntime.InferenceSession("recognitionModel.onnx")
ort_inputs = {ort_session.get_inputs()[0].name: to_numpy(image)}
ort_outs = ort_session.run(None, ort_inputs)
preds = torch.from_numpy(ort_outs[0])
```
Remember to `import onnxruntime` in the file header.
We can see how we are only passing one input entity. Although this model, in theory, is supposed to receive two. As with the detection model, the input must be transformed from a Tensor to a numpy array. We convert the output from an array to a Tensor, so that the data flow continues normally.
**To test, we can run EasyOCR with some image and see the result.**
## Others
We can use this function to compare the output of the PyTorch model and the ONNX model to quantify the difference:
`np.testing.assert_allclose(to_numpy(<PYTORCH_PREDICTION>), <ONNX_PREDICTION>, rtol=1e-03, atol=1e-05)`
In my tests, the difference between the detection models is minimal and passes the test correctly.
In case of the difference in the recognition models, the difference is slightly larger and the test fails. In spite of this it fails by very little and I have not observed failures in the actual recognition of the characters. I don't know if this is due to the problem with ONNX not detecting the two input entities, the problem with AdaptiveAvgPool2d or just a natural error in the model export and decimal approximations.
## Final note
I hope this will be of help to continue with the development of this excellent tool. I hope that exporters in EasyOCR and Pytorch can review this and find the answers to the questions raised.
|
open
|
2022-06-05T22:38:00Z
|
2025-03-05T11:58:29Z
|
https://github.com/JaidedAI/EasyOCR/issues/746
|
[] |
Kromtar
| 42
|
rthalley/dnspython
|
asyncio
| 599
|
tsigkey not recognized by peer with 2.0.0 (but works with 1.16.0)
|
Something changed in how tsigkey names are transmitted to the DNS server. This is the exact same code with 2.0.0 vs 1.16.0:
> (pyddns) mira/scanner (64) $ ./dnspython-delete-name.py usenet CNAME kamidake
> Deleting key 'usenet', of type 'CNAME' with value 'kamidake' in 'apricot.com'
> Traceback (most recent call last):
> File "./dnspython-delete-name.py", line 59, in <module>
> main()
> File "./dnspython-delete-name.py", line 50, in main
> response = dns.query.tcp(update, args['--dns_server'])
> File "/Users/scanner/.virtualenvs/pyddns/lib/python3.8/site-packages/dns/query.py", line 759, in tcp
> (r, received_time) = receive_tcp(s, expiration, one_rr_per_rrset,
> File "/Users/scanner/.virtualenvs/pyddns/lib/python3.8/site-packages/dns/query.py", line 691, in receive_tcp
> r = dns.message.from_wire(wire, keyring=keyring, request_mac=request_mac,
> File "/Users/scanner/.virtualenvs/pyddns/lib/python3.8/site-packages/dns/message.py", line 933, in from_wire
> m = reader.read()
> File "/Users/scanner/.virtualenvs/pyddns/lib/python3.8/site-packages/dns/message.py", line 859, in read
> self._get_section(MessageSection.ADDITIONAL, adcount)
> File "/Users/scanner/.virtualenvs/pyddns/lib/python3.8/site-packages/dns/message.py", line 820, in _get_section
> dns.tsig.validate(self.parser.wire,
> File "/Users/scanner/.virtualenvs/pyddns/lib/python3.8/site-packages/dns/tsig.py", line 183, in validate
> raise PeerBadKey
> dns.tsig.PeerBadKey: The peer didn't know the key we used
vs:
> (pyddns) mira/scanner (70) $ ./dnspython-delete-name.py usenet CNAME kamidake
> Deleting key 'usenet', of type 'CNAME' with value 'kamidake' in 'apricot.com'
> Got response: id 43936
> opcode UPDATE
> rcode NOERROR
> flags QR
> ;ZONE
> apricot.com. IN SOA
> ;PREREQ
> ;UPDATE
> ;ADDITIONAL
```
keyring = dns.tsigkeyring.from_text({TSIG_KEYNAME: tsig_key})
....
update = dns.update.Update(args['--zone'], keyring=keyring)
update.delete(args['<name>'], args['<type>'], args['<value>'])
response = dns.query.tcp(update, args['--dns_server'])
```
|
closed
|
2020-10-28T23:17:35Z
|
2021-10-21T06:28:38Z
|
https://github.com/rthalley/dnspython/issues/599
|
[] |
scanner
| 3
|
ansible/ansible
|
python
| 84,075
|
`ansible-test` host properties detection sometimes tracebacks in CI
|
### Summary
Specifically, this https://github.com/ansible/ansible/blob/f1f0d9bd5355de5b45b894a9adf649abb2f97df5/test/lib/ansible_test/_internal/docker_util.py#L327C31-L327C37 causes an `IndexError`, meaning that blocks is sometimes a list of 2 elements and not 3.
### Issue Type
Bug Report
### Component Name
ansible-test
### Ansible Version
```console
devel
```
### Configuration
```console
N/A
```
### OS / Environment
Ubuntu 24.04 in our CI
### Steps to Reproduce
Context: https://dev.azure.com/ansible/ansible/_build/results?buildId=125234&view=logs&j=6ce7cab1-69aa-56f4-11b1-869c767eb409&t=84972f9e-94d6-5fa2-afad-226d677b2f72&l=150
### Expected Results
Exception is handled, but I don't know what causes `ansible-test-probe` to print out less lines.
### Actual Results
```python-traceback
Traceback (most recent call last):
File "/__w/1/ansible/bin/ansible-test", line 44, in <module>
main()
File "/__w/1/ansible/bin/ansible-test", line 35, in main
cli_main(args)
File "/__w/1/ansible/test/lib/ansible_test/_internal/__init__.py", line 65, in main
main_internal(cli_args)
File "/__w/1/ansible/test/lib/ansible_test/_internal/__init__.py", line 91, in main_internal
args.func(config)
File "/__w/1/ansible/test/lib/ansible_test/_internal/commands/integration/posix.py", line 43, in command_posix_integration
host_state, internal_targets = command_integration_filter(args, all_targets)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/__w/1/ansible/test/lib/ansible_test/_internal/commands/integration/__init__.py", line 941, in command_integration_filter
cloud_init(args, internal_targets)
File "/__w/1/ansible/test/lib/ansible_test/_internal/commands/integration/cloud/__init__.py", line 162, in cloud_init
provider.setup()
File "/__w/1/ansible/test/lib/ansible_test/_internal/commands/integration/cloud/httptester.py", line 57, in setup
descriptor = run_support_container(
^^^^^^^^^^^^^^^^^^^^^^
File "/__w/1/ansible/test/lib/ansible_test/_internal/containers.py", line 157, in run_support_container
max_open_files = detect_host_properties(args).max_open_files
^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/__w/1/ansible/test/lib/ansible_test/_internal/docker_util.py", line 327, in detect_host_properties
mounts = MountEntry.loads(blocks[2])
~~~~~~^^^
IndexError: list index out of range
```
### Code of Conduct
- [X] I agree to follow the Ansible Code of Conduct
|
open
|
2024-10-08T13:34:04Z
|
2025-02-24T19:00:57Z
|
https://github.com/ansible/ansible/issues/84075
|
[
"bug",
"has_pr"
] |
webknjaz
| 14
|
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.