repo_name
stringlengths 9
75
| topic
stringclasses 30
values | issue_number
int64 1
203k
| title
stringlengths 1
976
| body
stringlengths 0
254k
| state
stringclasses 2
values | created_at
stringlengths 20
20
| updated_at
stringlengths 20
20
| url
stringlengths 38
105
| labels
listlengths 0
9
| user_login
stringlengths 1
39
| comments_count
int64 0
452
|
|---|---|---|---|---|---|---|---|---|---|---|---|
horovod/horovod
|
machine-learning
| 4,008
|
ipv6 address family
|
**Is your feature request related to a problem? Please describe.**
A clear and concise description of what the problem is. Ex. I'm always frustrated when [...]
I use horovod to launch discributed training tasks, and my cluster has been migirated to ipv6 family. However, the horovod.runner donot support the cluster with throwing errors, including but not limited to:
1. address parser at "parse_hosts_and_slots@common.utilhosts.py", the function requires ip addresses inputed are int the format of "IP:slot-nums", which conflcts with the ipv6 format.
2. server listen at "find_port@util.network.py", the addr=("",port) would generate a ipv4 addr.
3. BasicService@common.util.network.py, fail to support ipv6 family.
**Describe the solution you'd like**
A clear and concise description of what you want to happen.
current, I just add an environment to force the code execute with ipv6 branch, hoping a better solution from the official.
**Describe alternatives you've considered**
A clear and concise description of any alternative solutions or features you've considered.
**Additional context**
Add any other context or screenshots about the feature request here.
the horovod I used is based on pip3 install with a version=0.28.1
|
open
|
2023-12-11T04:16:15Z
|
2023-12-11T04:16:15Z
|
https://github.com/horovod/horovod/issues/4008
|
[
"enhancement"
] |
NEWPLAN
| 0
|
ResidentMario/geoplot
|
matplotlib
| 229
|
AttributeError: 'str' object has no attribute 'set_array' when calling kdeplot with legend=True
|
First, thanks for the great library.
I noticed an issue when trying to use the kdeplot function when also passing legend=True. It works fine when legend is not passed. Here is a code sample which is failing for me:
```python
import pandas as pd
import geopandas
import geoplot
df = pd.DataFrame(
[
[ 27.547044 , -82.533896 ],
[ 27.4853062, -82.5652603],
[ 27.4782766, -82.5937605],
[ 27.5018049, -82.6089842],
[ 27.476826 , -82.5190818],
[ 27.49207 , -82.5964237],
],
columns=['latitude', 'longitude']
)
gdf = geopandas.GeoDataFrame(
df, geometry=geopandas.points_from_xy(df.longitude, df.latitude)
)
gdf.crs = 4326
geoplot.kdeplot(gdf, shade=True, legend=True)
```
From that I get the following traceback:
```python
---------------------------------------------------------------------------
AttributeError Traceback (most recent call last)
<ipython-input-29-ba321c5cab0f> in <module>
17 )
18 gdf.crs = 4326
---> 19 geoplot.kdeplot(gdf, shade=True, legend=True)
~/.pyenv/versions/3.8.5/lib/python3.8/site-packages/geoplot/geoplot.py in kdeplot(df, projection, extent, figsize, ax, clip, shade_lowest, **kwargs)
1360 return ax
1361
-> 1362 plot = KDEPlot(
1363 df, projection=projection, extent=extent, figsize=figsize, ax=ax, clip=clip,
1364 shade_lowest=shade_lowest,
~/.pyenv/versions/3.8.5/lib/python3.8/site-packages/geoplot/geoplot.py in __init__(self, df, **kwargs)
1336 verify_input=False
1337 )
-> 1338 self.paint_legend(supports_hue=True, supports_scale=False, verify_input=False)
1339 self.paint_clip()
1340
~/.pyenv/versions/3.8.5/lib/python3.8/site-packages/geoplot/geoplot.py in paint_legend(self, supports_hue, supports_scale, verify_input, scale_multiplier)
354 )
355
--> 356 self.cmap.set_array(self.hue)
357 try:
358 plt.gcf().colorbar(self.cmap, ax=self.ax, **legend_kwargs)
AttributeError: 'str' object has no attribute 'set_array'
```
Here are the versions of Python and relevant packages:
```
Python: 3.8.5
OS: Ubuntu 20.04
geoplot==0.4.1
geopandas==0.8.1
seaborn==0.11.0
matplotlib==3.3.2
pandas==0.25.3
Cartopy==0.18.0
descartes==1.1.0
contextily==1.0.1
mapclassify==2.3.0
```
|
closed
|
2020-11-13T17:24:11Z
|
2022-02-27T19:40:17Z
|
https://github.com/ResidentMario/geoplot/issues/229
|
[
"bug"
] |
nickderobertis
| 3
|
mwaskom/seaborn
|
data-visualization
| 2,820
|
Logarithmic hist plot with multi='dodge': unequal bar width
|
Using a hist plot with logarithmic x-axis results in unequal bar width.
Fix PR is provided in #2819
|
closed
|
2022-05-25T07:05:56Z
|
2022-06-11T23:42:20Z
|
https://github.com/mwaskom/seaborn/issues/2820
|
[
"bug",
"mod:distributions"
] |
infosec-it-init
| 3
|
MilesCranmer/PySR
|
scikit-learn
| 213
|
[Code cleanup] make options more hierarchical
|
The current list of options is way too long to be understood by a user. I think a refactoring should be done where new objects are used to hierarchically define the parameters.
For example, rather than have 8 parameters passed flatly for the mutation weightings, you could have a single MutationWeights object that could be passed - and would have additional documentation on what the mutation weightings are.
I think it would make sense to start by writing down a hierarchical parameter grouping, and go from there.
|
open
|
2022-11-01T18:35:41Z
|
2023-04-20T06:05:51Z
|
https://github.com/MilesCranmer/PySR/issues/213
|
[
"documentation",
"priority: mid"
] |
MilesCranmer
| 2
|
jschneier/django-storages
|
django
| 513
|
Respect max_length when AWS_S3_FILE_OVERWRITE=True
|
`get_available_name` method of `S3Boto3Storage` doesn't respect `max_length`.
The Django Storage's `get_available_name` method truncates the file name if it's length is more than `max_length`.
When `AWS_S3_FILE_OVERWRITE=True` and len(file_name) > max_length, this results in Database error - `DataError: (1406, "Data too long for column 'file_field' at row 1")`.
```
def get_available_name(self, name, max_length=None):
"""Overwrite existing file with the same name."""
if self.file_overwrite:
name = self._clean_name(name)
>> ** CHANGE REQUIRED HERE: max_length VALIDATION **
return name
return super(S3Boto3Storage, self).get_available_name(name, max_length)
```
Validation for `max_length` should be added to the overridden method, when file_overwrite=True.
|
closed
|
2018-06-29T11:06:27Z
|
2018-08-30T21:59:54Z
|
https://github.com/jschneier/django-storages/issues/513
|
[
"bug",
"s3boto"
] |
a1Gupta
| 1
|
scikit-learn/scikit-learn
|
python
| 30,284
|
Create a process for releasing a wheel for a new Python version with a previous sklearn version on CI
|
For version 1.5.2, the wheels were not updated from the CI, but from an API key. Moving forward, I think we should update our CI to allow us to push specific python versions. I propose this process:
1. **Prerequisite**: Python 3.14rc support added to `cibuildwheel` + `numpy` & `scipy` has wheels for it
2. Update `build_tools/cirrus/arm_wheel.yml` and `.github/workflows/wheels.yml` to support the new version on `1.X.X` branch
3. Trigger `.github/workflows/publish_pypi.yml` (`workflow_run`) with a specific python version which will only upload wheels for that python version.
These are the tasks I see:
- **Required**: Update `.github/workflows/publish_pypi.yml` to accept a specific python version and only upload that python version.
- **Nice to have, but not required**: `build_tools/cirrus/arm_wheel.yml` and `.github/workflows/wheels.yml` to only build wheels for a specific python version.
CC @lesteve
|
open
|
2024-11-15T17:01:28Z
|
2024-11-28T15:35:00Z
|
https://github.com/scikit-learn/scikit-learn/issues/30284
|
[
"Build / CI"
] |
thomasjpfan
| 2
|
Johnserf-Seed/TikTokDownload
|
api
| 213
|
[BUG]无法抓取1080
|
按步骤编译后,只能抓到720p。但使用别的软件就可以抓到1080的视频 。
单独和批量都一样。
感谢作者的伟大贡献!
-操作系统:[例如windows10 64bit]
-vpn代理[例如开启、关闭]
-版本[如1.2.3]
|
closed
|
2022-09-08T05:51:31Z
|
2022-09-12T05:22:00Z
|
https://github.com/Johnserf-Seed/TikTokDownload/issues/213
|
[
"故障(bug)",
"额外求助(help wanted)",
"无效(invalid)"
] |
regdak
| 4
|
Asabeneh/30-Days-Of-Python
|
matplotlib
| 359
|
Translation Spanish
|
Hi. Thank you for your tutorials.
I want to add Spanish translation to this repository.
Can I work with that?
|
open
|
2023-02-20T18:34:10Z
|
2023-08-17T18:16:04Z
|
https://github.com/Asabeneh/30-Days-Of-Python/issues/359
|
[] |
Misio942
| 3
|
aio-libs/aiomysql
|
asyncio
| 720
|
Port PyMySQL changes
|
this is a very broad scope - probably needs a review of most aiomysql code to compare with pymysql code.
a good step to start should be porting all PyMySQL test cases to see which issues we can already identify from missing test cases.
an alternative could be to review all PyMySQL commits/changes between the previously used version and the current latest version: https://github.com/PyMySQL/PyMySQL/compare/v0.9.3...v1.0.2
this may however missing ports of older changes, I don't know how close attention was paid to this before.
|
open
|
2022-01-30T21:51:09Z
|
2022-05-10T16:15:43Z
|
https://github.com/aio-libs/aiomysql/issues/720
|
[
"pymysql",
"pymysql-port"
] |
Nothing4You
| 2
|
aiogram/aiogram
|
asyncio
| 825
|
Failed on aiogram.contrib.fsm_storage.redis.AioRedisAdapterV1.get
|
## Context
* Operating System: MacOS
* Python Version: 3.8.8
* aiogram version: 2.18
* aiohttp version: 3.8.1
## Expected Behavior
Trying to handle messages using template [AlbumMiddleware()
](https://github.com/shevasa/BotCustomDeclaration/blob/57eab3e5ce5c70b4d8f3cf4cd2aa73bb3e18a92f/middlewares/for_albums.py)
### Steps to Reproduce
Please provide detailed steps for reproducing the issue.
1. register AlbumMiddleware()
2. send media group to bot
probably "aiogram_dialog" also matter here
### Failure Logs
```python
2022-01-31 22:37:11,850 - asyncio - ERROR - Task exception was never retrieved
future: <Task finished name='Task-28' coro=<Dispatcher._process_polling_updates() done, defined at /Users/ismirnov/miniconda3/envs/bots/lib/python3.8/site-packages/aiogram/dispatcher/dispatcher.py:407> exception=AttributeError("'NoneType' object has no attribute 'get'")>
Traceback (most recent call last):
File "/Users/ismirnov/miniconda3/envs/bots/lib/python3.8/site-packages/aiogram/dispatcher/dispatcher.py", line 415, in _process_polling_updates
for responses in itertools.chain.from_iterable(await self.process_updates(updates, fast)):
File "/Users/ismirnov/miniconda3/envs/bots/lib/python3.8/site-packages/aiogram/dispatcher/dispatcher.py", line 235, in process_updates
return await asyncio.gather(*tasks)
File "/Users/ismirnov/miniconda3/envs/bots/lib/python3.8/site-packages/aiogram/dispatcher/handler.py", line 116, in notify
response = await handler_obj.handler(*args, **partial_data)
File "/Users/ismirnov/miniconda3/envs/bots/lib/python3.8/site-packages/aiogram/dispatcher/dispatcher.py", line 256, in process_update
return await self.message_handlers.notify(update.message)
File "/Users/ismirnov/miniconda3/envs/bots/lib/python3.8/site-packages/aiogram/dispatcher/handler.py", line 100, in notify
await self.dispatcher.middleware.trigger(f"pre_process_{self.middleware_key}", args + (data,))
File "/Users/ismirnov/miniconda3/envs/bots/lib/python3.8/site-packages/aiogram/dispatcher/middlewares.py", line 53, in trigger
await app.trigger(action, args)
File "/Users/ismirnov/miniconda3/envs/bots/lib/python3.8/site-packages/aiogram/dispatcher/middlewares.py", line 106, in trigger
await handler(*args)
File "/Users/ismirnov/miniconda3/envs/bots/lib/python3.8/site-packages/aiogram_dialog/context/intent_filter.py", line 56, in on_pre_process_message
stack = await proxy.load_stack()
File "/Users/ismirnov/miniconda3/envs/bots/lib/python3.8/site-packages/aiogram_dialog/context/storage.py", line 32, in load_stack
data = await self.storage.get_data(
File "/Users/ismirnov/miniconda3/envs/bots/lib/python3.8/site-packages/aiogram/contrib/fsm_storage/redis.py", line 446, in get_data
raw_result = await redis.get(key)
File "/Users/ismirnov/miniconda3/envs/bots/lib/python3.8/site-packages/aiogram/contrib/fsm_storage/redis.py", line 310, in get
return await self._redis.get(name, encoding="utf8", **kwargs)
AttributeError: 'NoneType' object has no attribute 'get'
```
Fixed with changing aiogram.contrib.fsm_storage.redis.AioRedisAdapterV1.get
```python
async def get(self, name, **kwargs):
if not self._redis:
await self.get_redis()
return await self._redis.get(name, encoding="utf8", **kwargs)
```
any ideas or should I prepare a PR?
|
closed
|
2022-02-01T14:13:30Z
|
2023-08-04T18:21:30Z
|
https://github.com/aiogram/aiogram/issues/825
|
[
"needs triage"
] |
brewbytes-dev
| 4
|
nerfstudio-project/nerfstudio
|
computer-vision
| 2,901
|
ModuleNotFoundError: No module named 'nerfstudio.viewer.viewer_elements'
|
Hi,
Thank you for sharing the code.
I don't have the problem before when using nerfstudio but this week I am having this error: 'ModuleNotFoundError: No module named 'nerfstudio.viewer.viewer_elements'', although using the same Docker as before.
Do you have any clue about this problem?
Thanks!
|
open
|
2024-02-11T05:31:02Z
|
2024-04-29T08:24:31Z
|
https://github.com/nerfstudio-project/nerfstudio/issues/2901
|
[] |
br0202
| 2
|
httpie/cli
|
api
| 811
|
Http headers colors should be based on status code only
|
As I commented in #735, some servers (eg Tomcat) don't return the reason-phrase, and it seems valid not to ([RFC 7230](https://tools.ietf.org/html/rfc7230#page-22)).
So "200" (instead of "200 OK") should not render headers in red, I think.
|
closed
|
2019-10-29T16:42:47Z
|
2021-02-18T23:40:08Z
|
https://github.com/httpie/cli/issues/811
|
[] |
Guillaume-Mayer
| 4
|
alteryx/featuretools
|
data-science
| 2,464
|
release Featuretools v1.22.0
|
- release instructions: https://github.com/alteryx/featuretools/blob/main/release.md
|
closed
|
2023-01-25T18:30:54Z
|
2023-02-01T15:24:35Z
|
https://github.com/alteryx/featuretools/issues/2464
|
[] |
gsheni
| 0
|
tqdm/tqdm
|
jupyter
| 1,283
|
`ipywidgets` variant broken
|
```python
import sys, time, tqdm
for j in tqdm.trange(100, file=sys.stdout, leave=False, unit_scale=True, desc='loop'):
time.sleep(1)
```
works, but
```python
for j in tqdm.auto.tqdm(range(100), file=sys.stdout, leave=False, unit_scale=True, desc='loop'):
time.sleep(1)
````
shows a frozen progress bar and no percent update:
```
loop: 0%| | 0.00/100 [00:00<?, ?it/s]
```
<details><summary><b>conda list</b></summary>
```
# packages in environment at D:\Anaconda\envs\pyt:
#
# Name Version Build Channel
absl-py 0.15.0 pyhd8ed1ab_0 conda-forge
aiohttp 3.7.4.post0 py38h294d835_1 conda-forge
alabaster 0.7.12 py_0 conda-forge
anyio 3.3.3 py38haa244fe_0 conda-forge
appdirs 1.4.4 pyh9f0ad1d_0 conda-forge
argh 0.26.2 pyh9f0ad1d_1002 conda-forge
argon2-cffi 21.1.0 py38h294d835_0 conda-forge
arrow 1.2.0 pyhd8ed1ab_0 conda-forge
astroid 2.5.8 py38haa244fe_0 conda-forge
async-timeout 3.0.1 py_1000 conda-forge
async_generator 1.10 py_0 conda-forge
atomicwrites 1.4.0 pyh9f0ad1d_0 conda-forge
attrs 21.2.0 pyhd8ed1ab_0 conda-forge
audioread 2.1.9 py38haa244fe_0 conda-forge
autopep8 1.6.0 pyhd8ed1ab_1 conda-forge
babel 2.9.1 pyh44b312d_0 conda-forge
backcall 0.2.0 pyh9f0ad1d_0 conda-forge
backports 1.0 py_2 conda-forge
backports.functools_lru_cache 1.6.4 pyhd8ed1ab_0 conda-forge
bcrypt 3.2.0 py38h294d835_1 conda-forge
binaryornot 0.4.4 py_1 conda-forge
black 21.9b0 pyhd8ed1ab_0 conda-forge
blas 1.0 mkl
bleach 4.1.0 pyhd8ed1ab_0 conda-forge
blinker 1.4 py_1 conda-forge
brotli-python 1.0.9 py38h885f38d_5 conda-forge
brotlipy 0.7.0 py38h294d835_1001 conda-forge
bzip2 1.0.8 h8ffe710_4 conda-forge
ca-certificates 2021.10.26 haa95532_2
cached-property 1.5.2 hd8ed1ab_1 conda-forge
cached_property 1.5.2 pyha770c72_1 conda-forge
cachetools 4.2.4 pyhd8ed1ab_0 conda-forge
certifi 2021.10.8 py38haa244fe_1 conda-forge
cffi 1.14.6 py38hd8c33c5_1 conda-forge
chardet 4.0.0 py38haa244fe_1 conda-forge
charset-normalizer 2.0.0 pyhd8ed1ab_0 conda-forge
click 7.1.2 pyh9f0ad1d_0 conda-forge
cloudpickle 2.0.0 pyhd8ed1ab_0 conda-forge
colorama 0.4.4 pyh9f0ad1d_0 conda-forge
conda 4.11.0 py38haa244fe_0 conda-forge
conda-package-handling 1.7.3 py38h31c79cd_1 conda-forge
configparser 5.1.0 pyhd8ed1ab_0 conda-forge
cookiecutter 1.6.0 py38_1000 conda-forge
cryptography 3.4.7 py38hd7da0ea_0 conda-forge
cudatoolkit 11.3.1 h59b6b97_2
cupy 9.5.0 py38hf95616d_1 conda-forge
cycler 0.10.0 py_2 conda-forge
cython 0.29.24 py38h885f38d_0 conda-forge
dash 2.0.0 pyhd8ed1ab_0 conda-forge
dataclasses 0.8 pyhc8e2a94_3 conda-forge
debugpy 1.4.1 py38h885f38d_0 conda-forge
decorator 5.1.0 pyhd8ed1ab_0 conda-forge
defusedxml 0.7.1 pyhd8ed1ab_0 conda-forge
diff-match-patch 20200713 pyh9f0ad1d_0 conda-forge
docker-pycreds 0.4.0 py_0 conda-forge
docutils 0.17.1 py38haa244fe_0 conda-forge
entrypoints 0.3 pyhd8ed1ab_1003 conda-forge
fastrlock 0.8 py38h885f38d_1 conda-forge
fftw 3.3.10 nompi_hea9a5d6_101 conda-forge
flake8 4.0.1 pyhd8ed1ab_1 conda-forge
flask 2.0.2 pyhd8ed1ab_0 conda-forge
flask-compress 1.10.1 pyhd8ed1ab_0 conda-forge
freetype 2.10.4 h546665d_1 conda-forge
fsspec 2021.10.1 pyhd8ed1ab_0 conda-forge
future 0.18.2 py38haa244fe_3 conda-forge
gitdb 4.0.9 pyhd8ed1ab_0 conda-forge
gitpython 3.1.24 pyhd8ed1ab_0 conda-forge
google-auth 1.35.0 pyh6c4a22f_0 conda-forge
google-auth-oauthlib 0.4.6 pyhd8ed1ab_0 conda-forge
grpcio 1.41.1 py38he5377a8_1 conda-forge
h5py 3.6.0 nompi_py38hde0384b_100 conda-forge
hdf5 1.12.1 nompi_h2a0e4a3_103 conda-forge
icu 68.1 h0e60522_0 conda-forge
idna 3.1 pyhd3deb0d_0 conda-forge
imagesize 1.2.0 py_0 conda-forge
importlib-metadata 4.2.0 py38haa244fe_0 conda-forge
importlib_metadata 4.2.0 hd8ed1ab_0 conda-forge
inflection 0.5.1 pyh9f0ad1d_0 conda-forge
iniconfig 1.1.1 pyh9f0ad1d_0 conda-forge
intel-openmp 2021.3.0 h57928b3_3372 conda-forge
intervaltree 3.0.2 py_0 conda-forge
ipykernel 6.4.1 py38h595d716_0 conda-forge
ipython 7.28.0 py38h595d716_0 conda-forge
ipython_genutils 0.2.0 py_1 conda-forge
ipywidgets 7.6.5 pyhd8ed1ab_0 conda-forge
isort 5.9.3 pyhd8ed1ab_0 conda-forge
itsdangerous 2.0.1 pyhd8ed1ab_0 conda-forge
jbig 2.1 h8d14728_2003 conda-forge
jedi 0.18.0 py38haa244fe_2 conda-forge
jellyfish 0.8.9 py38h294d835_2 conda-forge
jinja2 3.0.2 pyhd8ed1ab_0 conda-forge
jinja2-time 0.2.0 py_2 conda-forge
joblib 1.1.0 pyhd8ed1ab_0 conda-forge
jpeg 9d h8ffe710_0 conda-forge
json5 0.9.6 pyhd3eb1b0_0
jsonschema 4.1.0 pyhd8ed1ab_0 conda-forge
jupyter-console 6.4.0 pypi_0 pypi
jupyter_client 6.1.12 pyhd8ed1ab_0 conda-forge
jupyter_core 4.8.1 py38haa244fe_0 conda-forge
jupyter_server 1.11.1 pyhd8ed1ab_0 conda-forge
jupyterlab 3.2.1 pyhd8ed1ab_0 conda-forge
jupyterlab-server 1.2.0 pypi_0 pypi
jupyterlab_pygments 0.1.2 pyh9f0ad1d_0 conda-forge
jupyterlab_server 2.8.2 pyhd8ed1ab_0 conda-forge
jupyterlab_widgets 1.0.2 pyhd8ed1ab_0 conda-forge
keyring 23.2.1 py38haa244fe_0 conda-forge
kiwisolver 1.3.2 py38hbd9d945_0 conda-forge
krb5 1.19.2 h20d022d_3 conda-forge
lazy-object-proxy 1.6.0 py38h294d835_0 conda-forge
lcms2 2.12 h2a16943_0 conda-forge
lerc 2.2.1 h0e60522_0 conda-forge
libarchive 3.5.2 hb45042f_1 conda-forge
libblas 3.9.0 11_win64_mkl conda-forge
libcblas 3.9.0 11_win64_mkl conda-forge
libclang 11.1.0 default_h5c34c98_1 conda-forge
libcurl 7.80.0 h789b8ee_1 conda-forge
libdeflate 1.7 h8ffe710_5 conda-forge
libflac 1.3.3 h0e60522_1 conda-forge
libiconv 1.16 he774522_0 conda-forge
liblapack 3.9.0 11_win64_mkl conda-forge
libmamba 0.19.1 h44daa3b_0 conda-forge
libmambapy 0.19.1 py38h2bfd5b9_0 conda-forge
libogg 1.3.5 h2bbff1b_1
libopus 1.3.1 h8ffe710_1 conda-forge
libpng 1.6.37 h1d00b33_2 conda-forge
libprotobuf 3.19.1 h7755175_0 conda-forge
librosa 0.8.1 pyhd8ed1ab_0 conda-forge
libsndfile 1.0.31 h0e60522_1 conda-forge
libsodium 1.0.18 h8d14728_1 conda-forge
libsolv 0.7.19 h7755175_5 conda-forge
libspatialindex 1.9.3 h39d44d4_4 conda-forge
libssh2 1.10.0 h680486a_2 conda-forge
libtiff 4.3.0 h0c97f57_1 conda-forge
libuv 1.40.0 he774522_0
libvorbis 1.3.7 ha925a31_0 conda-forge
libxml2 2.9.12 hf5bbc77_1 conda-forge
libzlib 1.2.11 h8ffe710_1013 conda-forge
llvmlite 0.36.0 py38h57a6900_0 conda-forge
lz4-c 1.9.3 h8ffe710_1 conda-forge
lzo 2.10 hfa6e2cd_1000 conda-forge
m2w64-gcc-libgfortran 5.3.0 6 conda-forge
m2w64-gcc-libs 5.3.0 7 conda-forge
m2w64-gcc-libs-core 5.3.0 7 conda-forge
m2w64-gmp 6.1.0 2 conda-forge
m2w64-libwinpthread-git 5.0.0.4634.697f757 2 conda-forge
mamba 0.19.1 py38hecfeebb_0 conda-forge
markdown 3.3.4 pyhd8ed1ab_0 conda-forge
markupsafe 2.0.1 py38h294d835_0 conda-forge
matplotlib 3.4.3 py38haa244fe_1 conda-forge
matplotlib-base 3.4.3 py38h1f000d6_1 conda-forge
matplotlib-inline 0.1.3 pyhd8ed1ab_0 conda-forge
mccabe 0.6.1 py_1 conda-forge
menuinst 1.4.18 py38haa244fe_1 conda-forge
mistune 0.8.4 py38h294d835_1004 conda-forge
mkl 2021.3.0 hb70f87d_564 conda-forge
more-itertools 8.10.0 pyhd8ed1ab_0 conda-forge
mpmath 1.2.1 pyhd8ed1ab_0 conda-forge
msys2-conda-epoch 20160418 1 conda-forge
multidict 5.2.0 py38h294d835_1 conda-forge
mypy_extensions 0.4.3 py38haa244fe_3 conda-forge
nbclassic 0.3.2 pyhd8ed1ab_0 conda-forge
nbclient 0.5.4 pyhd8ed1ab_0 conda-forge
nbconvert 5.6.1 pypi_0 pypi
nbformat 5.1.3 pyhd8ed1ab_0 conda-forge
nest-asyncio 1.5.1 pyhd8ed1ab_0 conda-forge
ninja 1.10.2 h6d14046_1
notebook 6.4.4 pyha770c72_0 conda-forge
numba 0.53.0 py38h5c177ec_0 conda-forge
numpy 1.21.2 py38h089cfbf_0 conda-forge
numpydoc 1.1.0 py_1 conda-forge
oauthlib 3.1.1 pyhd8ed1ab_0 conda-forge
olefile 0.46 pyh9f0ad1d_1 conda-forge
openjpeg 2.4.0 hb211442_1 conda-forge
openssl 1.1.1l h8ffe710_0 conda-forge
packaging 21.0 pyhd8ed1ab_0 conda-forge
pandas 1.3.3 py38h5d928e2_0 conda-forge
pandoc 2.14.2 h8ffe710_0 conda-forge
pandocfilters 1.5.0 pyhd8ed1ab_0 conda-forge
paramiko 2.7.2 pyh9f0ad1d_0 conda-forge
parso 0.8.2 pyhd8ed1ab_0 conda-forge
pathspec 0.9.0 pyhd8ed1ab_0 conda-forge
pathtools 0.1.2 py_1 conda-forge
pdfkit 0.6.1 pypi_0 pypi
pexpect 4.8.0 pyh9f0ad1d_2 conda-forge
pickleshare 0.7.5 py_1003 conda-forge
pillow 8.3.2 py38h794f750_0 conda-forge
pip 21.2.4 pyhd8ed1ab_0 conda-forge
platformdirs 2.3.0 pyhd8ed1ab_0 conda-forge
plotly 5.3.1 py_0 plotly
pluggy 1.0.0 py38haa244fe_1 conda-forge
pooch 1.5.2 pyhd8ed1ab_0 conda-forge
poyo 0.5.0 py_0 conda-forge
prometheus_client 0.11.0 pyhd8ed1ab_0 conda-forge
promise 2.3 py38haa244fe_5 conda-forge
prompt-toolkit 3.0.20 pyha770c72_0 conda-forge
protobuf 3.19.1 py38h885f38d_1 conda-forge
psutil 5.8.0 py38h294d835_1 conda-forge
ptyprocess 0.7.0 pyhd3deb0d_0 conda-forge
py 1.10.0 pyhd3deb0d_0 conda-forge
py-lz4framed 0.14.0 pypi_0 pypi
pyasn1 0.4.8 py_0 conda-forge
pyasn1-modules 0.2.8 py_0
pybind11-abi 4 hd8ed1ab_3 conda-forge
pycodestyle 2.8.0 pyhd8ed1ab_0 conda-forge
pycosat 0.6.3 py38h294d835_1009 conda-forge
pycparser 2.20 pyh9f0ad1d_2 conda-forge
pydeprecate 0.3.1 pyhd8ed1ab_0 conda-forge
pydocstyle 6.1.1 pyhd8ed1ab_0 conda-forge
pyfftw 0.12.0 py38h46b76f8_3 conda-forge
pyflakes 2.4.0 pyhd8ed1ab_0 conda-forge
pygments 2.10.0 pyhd8ed1ab_0 conda-forge
pyjwt 2.3.0 pyhd8ed1ab_0 conda-forge
pylint 2.7.2 py38haa244fe_0 conda-forge
pyls-spyder 0.4.0 pyhd8ed1ab_0 conda-forge
pynacl 1.4.0 py38h31c79cd_2 conda-forge
pyopenssl 21.0.0 pyhd8ed1ab_0 conda-forge
pyparsing 2.4.7 pyh9f0ad1d_0 conda-forge
pypiwin32 223 pypi_0 pypi
pyqt 5.12.3 py38haa244fe_7 conda-forge
pyqt-impl 5.12.3 py38h885f38d_7 conda-forge
pyqt5-sip 4.19.18 py38h885f38d_7 conda-forge
pyqtchart 5.12 py38h885f38d_7 conda-forge
pyqtwebengine 5.12.1 py38h885f38d_7 conda-forge
pyrsistent 0.17.3 py38h294d835_2 conda-forge
pysocks 1.7.1 py38haa244fe_3 conda-forge
pysoundfile 0.10.3.post1 pyhd3deb0d_0 conda-forge
pytest 6.2.5 py38haa244fe_0 conda-forge
python 3.8.12 h7840368_1_cpython conda-forge
python-dateutil 2.8.2 pyhd8ed1ab_0 conda-forge
python-lsp-black 1.0.0 pyhd8ed1ab_0 conda-forge
python-lsp-jsonrpc 1.0.0 pyhd8ed1ab_0 conda-forge
python-lsp-server 1.3.3 pyhd8ed1ab_0 conda-forge
python_abi 3.8 2_cp38 conda-forge
pytorch 1.10.0 py3.8_cuda11.3_cudnn8_0 pytorch
pytorch-lightning 1.5.6 pyhd8ed1ab_0 conda-forge
pytorch-mutex 1.0 cuda pytorch
pytz 2021.3 pyhd8ed1ab_0 conda-forge
pyu2f 0.1.5 pyhd8ed1ab_0 conda-forge
pywin32 301 py38h294d835_0 conda-forge
pywin32-ctypes 0.2.0 py38haa244fe_1003 conda-forge
pywinpty 1.1.4 py38hd3f51b4_0 conda-forge
pyyaml 5.4.1 py38h294d835_1 conda-forge
pyzmq 22.3.0 py38h09162b1_0 conda-forge
qdarkstyle 3.0.2 pyhd8ed1ab_0 conda-forge
qstylizer 0.2.1 pyhd8ed1ab_0 conda-forge
qt 5.12.9 h5909a2a_4 conda-forge
qtawesome 1.0.3 pyhd8ed1ab_0 conda-forge
qtconsole 5.2.2 pyhd8ed1ab_0 conda-forge
qtpy 1.11.2 pyhd8ed1ab_0 conda-forge
regex 2021.10.8 py38h294d835_0 conda-forge
reproc 14.2.3 h8ffe710_0 conda-forge
reproc-cpp 14.2.3 h0e60522_0 conda-forge
requests 2.26.0 pyhd8ed1ab_0 conda-forge
requests-oauthlib 1.3.0 pyh9f0ad1d_0 conda-forge
requests-unixsocket 0.2.0 py_0 conda-forge
resampy 0.2.2 py_0 conda-forge
rope 0.20.1 pyhd8ed1ab_0 conda-forge
rsa 4.7.2 pyh44b312d_0 conda-forge
rtree 0.9.7 py38h8b54edf_2 conda-forge
ruamel_yaml 0.15.100 py38h2bbff1b_0
scikit-learn 1.0 py38h8224a6f_1 conda-forge
scipy 1.7.1 py38ha1292f7_0 conda-forge
send2trash 1.8.0 pyhd8ed1ab_0 conda-forge
sentry-sdk 1.5.0 pyhd8ed1ab_0 conda-forge
setuptools 58.2.0 py38haa244fe_0 conda-forge
shortuuid 1.0.8 py38haa244fe_0 conda-forge
six 1.16.0 pyh6c4a22f_0 conda-forge
smmap 3.0.5 pyh44b312d_0 conda-forge
sniffio 1.2.0 py38haa244fe_1 conda-forge
snowballstemmer 2.1.0 pyhd8ed1ab_0 conda-forge
sortedcontainers 2.4.0 pyhd8ed1ab_0 conda-forge
sounddevice 0.4.3 pypi_0 pypi
sphinx 4.2.0 pyh6c4a22f_0 conda-forge
sphinxcontrib-applehelp 1.0.2 py_0 conda-forge
sphinxcontrib-devhelp 1.0.2 py_0 conda-forge
sphinxcontrib-htmlhelp 2.0.0 pyhd8ed1ab_0 conda-forge
sphinxcontrib-jsmath 1.0.1 py_0 conda-forge
sphinxcontrib-qthelp 1.0.3 py_0 conda-forge
sphinxcontrib-serializinghtml 1.1.5 pyhd8ed1ab_0 conda-forge
spyder 5.2.1 py38haa244fe_0 conda-forge
spyder-kernels 2.2.0 py38haa244fe_0 conda-forge
sqlite 3.36.0 h8ffe710_2 conda-forge
subprocess32 3.5.4 py_1 conda-forge
sympy 1.9 py38haa244fe_0 conda-forge
tbb 2021.3.0 h2d74725_0 conda-forge
tenacity 8.0.1 py38haa95532_0
tensorboard 2.6.0 pyhd8ed1ab_1 conda-forge
tensorboard-data-server 0.6.0 py38haa244fe_1 conda-forge
tensorboard-plugin-wit 1.8.0 pyh44b312d_0 conda-forge
termcolor 1.1.0 py_2 conda-forge
terminado 0.12.1 py38haa244fe_0 conda-forge
testpath 0.5.0 pyhd8ed1ab_0 conda-forge
textdistance 4.2.1 pyhd8ed1ab_0 conda-forge
threadpoolctl 3.0.0 pyh8a188c0_0 conda-forge
three-merge 0.1.1 pyh9f0ad1d_0 conda-forge
tinycss2 1.1.0 pyhd8ed1ab_0 conda-forge
tk 8.6.11 h8ffe710_1 conda-forge
toml 0.10.2 pyhd8ed1ab_0 conda-forge
tomli 1.2.1 pyhd8ed1ab_0 conda-forge
torchinfo 1.5.4 pyhd8ed1ab_0 conda-forge
torchmetrics 0.6.0 pyhd8ed1ab_0 conda-forge
torchsummary 1.5.1 pypi_0 pypi
torchvision 0.11.1 py38_cu113 pytorch
tornado 6.1 py38h294d835_1 conda-forge
tqdm 4.62.3 pyhd8ed1ab_0 conda-forge
traitlets 4.3.3 pypi_0 pypi
typed-ast 1.4.3 py38h294d835_0 conda-forge
typing-extensions 3.10.0.2 hd8ed1ab_0 conda-forge
typing_extensions 3.10.0.2 pyha770c72_0 conda-forge
ucrt 10.0.20348.0 h57928b3_0 conda-forge
ujson 4.2.0 py38h885f38d_0 conda-forge
urllib3 1.26.7 pyhd8ed1ab_0 conda-forge
vc 14.2 hb210afc_5 conda-forge
vs2015_runtime 14.29.30037 h902a5da_5 conda-forge
wandb 0.12.9 pyhd8ed1ab_0 conda-forge
watchdog 2.1.6 py38haa244fe_0 conda-forge
wcwidth 0.2.5 pyh9f0ad1d_2 conda-forge
webencodings 0.5.1 py_1 conda-forge
websocket-client 0.58.0 py38haa95532_4
werkzeug 2.0.1 pyhd8ed1ab_0 conda-forge
wheel 0.37.0 pyhd8ed1ab_1 conda-forge
whichcraft 0.6.1 py_0 conda-forge
widgetsnbextension 3.5.2 py38haa244fe_0 conda-forge
win10toast 0.9 pypi_0 pypi
win_inet_pton 1.1.0 py38haa244fe_2 conda-forge
winpty 0.4.3 4 conda-forge
wrapt 1.12.1 py38h294d835_3 conda-forge
xz 5.2.5 h62dcd97_1 conda-forge
yaml 0.2.5 he774522_0 conda-forge
yaml-cpp 0.6.3 ha925a31_4 conda-forge
yapf 0.31.0 pyhd8ed1ab_0 conda-forge
yarl 1.7.2 py38h294d835_1 conda-forge
yaspin 2.1.0 pyhd8ed1ab_0 conda-forge
zeromq 4.3.4 h0e60522_1 conda-forge
zipp 3.6.0 pyhd8ed1ab_0 conda-forge
zlib 1.2.11 h8ffe710_1013 conda-forge
zstd 1.5.0 h6255e5f_0 conda-forge
```
</details>
<details><summary><b>conda info</b></summary>
```
active environment : pyt
active env location : D:\Anaconda\envs\pyt
shell level : 2
user config file : C:\Users\OverL\.condarc
populated config files : C:\Users\OverL\.condarc
conda version : 4.10.3
conda-build version : 3.18.11
python version : 3.8.3.final.0
virtual packages : __cuda=11.5=0
__win=0=0
__archspec=1=x86_64
base environment : D:\Anaconda (writable)
conda av data dir : D:\Anaconda\etc\conda
conda av metadata url : None
channel URLs : https://repo.anaconda.com/pkgs/main/win-64
https://repo.anaconda.com/pkgs/main/noarch
https://repo.anaconda.com/pkgs/r/win-64
https://repo.anaconda.com/pkgs/r/noarch
https://repo.anaconda.com/pkgs/msys2/win-64
https://repo.anaconda.com/pkgs/msys2/noarch
package cache : D:\Anaconda\pkgs
C:\Users\OverL\.conda\pkgs
C:\Users\OverL\AppData\Local\conda\conda\pkgs
envs directories : D:\Anaconda\envs
C:\Users\OverL\.conda\envs
C:\Users\OverL\AppData\Local\conda\conda\envs
platform : win-64
user-agent : conda/4.10.3 requests/2.24.0 CPython/3.8.3 Windows/10 Windows/10.0.19041
administrator : False
netrc file : C:\Users\OverL/.netrc
offline mode : False
```
</details>
Discovered in [PL](https://github.com/PyTorchLightning/pytorch-lightning/issues/11208)
|
open
|
2021-12-22T22:20:23Z
|
2022-09-14T15:03:40Z
|
https://github.com/tqdm/tqdm/issues/1283
|
[
"invalid ⛔",
"need-feedback 📢",
"p2-bug-warning ⚠",
"submodule-notebook 📓"
] |
OverLordGoldDragon
| 5
|
huggingface/datasets
|
computer-vision
| 6,465
|
`load_dataset` uses out-of-date cache instead of re-downloading a changed dataset
|
### Describe the bug
When a dataset is updated on the hub, using `load_dataset` will load the locally cached dataset instead of re-downloading the updated dataset
### Steps to reproduce the bug
Here is a minimal example script to
1. create an initial dataset and upload
2. download it so it is stored in cache
3. change the dataset and re-upload
4. redownload
```python
import time
from datasets import Dataset, DatasetDict, DownloadMode, load_dataset
username = "YOUR_USERNAME_HERE"
initial = Dataset.from_dict({"foo": [1, 2, 3]})
print(f"Intial {initial['foo']}")
initial_ds = DatasetDict({"train": initial})
initial_ds.push_to_hub("test")
time.sleep(1)
download = load_dataset(f"{username}/test", split="train")
changed = download.map(lambda x: {"foo": x["foo"] + 1})
print(f"Changed {changed['foo']}")
changed.push_to_hub("test")
time.sleep(1)
download_again = load_dataset(f"{username}/test", split="train")
print(f"Download Changed {download_again['foo']}")
# >>> gives the out-dated [1,2,3] when it should be changed [2,3,4]
```
The redownloaded dataset should be the changed dataset but it is actually the cached, initial dataset. Force-redownloading gives the correct dataset
```python
download_again_force = load_dataset(f"{username}/test", split="train", download_mode=DownloadMode.FORCE_REDOWNLOAD)
print(f"Force Download Changed {download_again_force['foo']}")
# >>> [2,3,4]
```
### Expected behavior
I assumed there should be some sort of hashing that should check for changes in the dataset and re-download if the hashes don't match
### Environment info
- `datasets` version: 2.15.0 │
- Platform: Linux-5.15.0-1028-nvidia-x86_64-with-glibc2.17 │
- Python version: 3.8.17 │
- `huggingface_hub` version: 0.19.4 │
- PyArrow version: 13.0.0 │
- Pandas version: 2.0.3 │
- `fsspec` version: 2023.6.0
|
open
|
2023-12-02T21:35:17Z
|
2024-08-20T08:32:11Z
|
https://github.com/huggingface/datasets/issues/6465
|
[] |
mnoukhov
| 2
|
xinntao/Real-ESRGAN
|
pytorch
| 33
|
x2 doesn't working at all
|

But anyway this is the best upscale tool I ever seen before
|
closed
|
2021-08-15T16:50:53Z
|
2021-08-19T02:21:08Z
|
https://github.com/xinntao/Real-ESRGAN/issues/33
|
[] |
crwg
| 13
|
huggingface/datasets
|
pytorch
| 7,364
|
API endpoints for gated dataset access requests
|
### Feature request
I would like a programatic way of requesting access to gated datasets. The current solution to gain access forces me to visit a website and physically click an "agreement" button (as per the [documentation](https://huggingface.co/docs/hub/en/datasets-gated#access-gated-datasets-as-a-user)).
An ideal approach would be HF API download methods that negotiate access on my behalf based on information from my CLI login and/or token. I realise that may be naive given the various types of access semantics available to dataset authors (automatic versus manual approval, for example) and complexities it might add to existing methods, but something along those lines would be nice.
Perhaps using the `*_access_request` methods available to dataset authors can be a precedent; see [`reject_access_request`](https://huggingface.co/docs/huggingface_hub/main/en/package_reference/hf_api#huggingface_hub.HfApi.reject_access_request) for example.
### Motivation
When trying to download files from a gated dataset, I'm met with a `GatedRepoError` and instructed to visit the repository's website to gain access:
```
Cannot access gated repo for url https://huggingface.co/datasets/open-llm-leaderboard/meta-llama__Meta-Llama-3.1-70B-Instruct-details/resolve/main/meta-llama__Meta-Llama-3.1-70B-Instruct/samples_leaderboard_math_precalculus_hard_2024-07-19T18-47-29.522341.jsonl.
Access to dataset open-llm-leaderboard/meta-llama__Meta-Llama-3.1-70B-Instruct-details is restricted and you are not in the authorized list. Visit https://huggingface.co/datasets/open-llm-leaderboard/meta-llama__Meta-Llama-3.1-70B-Instruct-details to ask for access.
```
This makes task automation extremely difficult. For example, I'm interested in studying sample-level responses of models on the LLM leaderboard -- how they answered particular questions on a given evaluation framework. As I come across more and more participants that gate their data, it's becoming unwieldy to continue my work (there over 2,000 participants, so in the worst case that's the number of website visits I'd need to manually undertake).
One approach is use Selenium to react to the `GatedRepoError`, but that seems like overkill; and a potential violation HF terms of service (?).
As mentioned in the previous section, there seems to be an [API for gated dataset owners](https://huggingface.co/docs/hub/en/datasets-gated#via-the-api) to managed access requests, and thus some appetite for allowing automated management of gating. This feature request is to extend that to dataset users.
### Your contribution
Whether I can help depends on a few things; one being the complexity of the underlying gated access design. If this feature request is accepted I am open to being involved in discussions and testing, and even development under the right time-outcome tradeoff.
|
closed
|
2025-01-09T06:21:20Z
|
2025-01-09T11:17:40Z
|
https://github.com/huggingface/datasets/issues/7364
|
[
"enhancement"
] |
jerome-white
| 3
|
arnaudmiribel/streamlit-extras
|
streamlit
| 81
|
Faker is super slow to load datasets
|
Maybe better to just use a same set of datasets throughout all extras and have them locally in the repo, instead of fetching all around
|
open
|
2022-11-18T15:39:01Z
|
2023-02-01T14:09:44Z
|
https://github.com/arnaudmiribel/streamlit-extras/issues/81
|
[
"enhancement"
] |
arnaudmiribel
| 0
|
dmlc/gluon-cv
|
computer-vision
| 1,708
|
It is possible to execute Alpha Pose Estimation using GPU?
|
I'm trying to execute the Alpha Pose estimation using GPU, but the following error is return:
`RuntimeError: Parameter 'conv0_weight' was not initialized on context cpu(0). It was only initialized on [gpu(0)].`
How can I fix this error? Or to run the Alpha Pose model using GPU do I need to run in another way?
Here it is a piece of my code:
import mxnet as mx
import gluoncv as gcv
from gluoncv import data
from gluoncv.model_zoo import get_model
from gluoncv.data.transforms.pose import detector_to_alpha_pose, heatmap_to_coord_alpha_pose
from gluoncv.utils.viz import cv_plot_image, cv_plot_keypoints
ctx = mx.gpu(0)
detector = get_model(''yolo3_mobilenet1.0_coco'', pretrained=True, ctx=ctx)
detector.reset_class(classes=['person'], reuse_weights=['person'])
net = get_model('alpha_pose_resnet101_v1b_coco', pretrained=True, ctx=ctx)
cap = cv2.VideoCapture('video.mp4')
fps = cap.get(cv2.CAP_PROP_FPS)
success, frame = cap.read()
height, width, _ = frame.shape
fourcc = cv2.VideoWriter_fourcc(*'XVID')
result = cv2.VideoWriter('output_filename.mp4',fourcc, fps, (width, height))
while True:
success, frame = cap.read()
if success:
frame = mx.nd.array(cv2.cvtColor(frame, cv2.COLOR_BGR2RGB)).astype('uint8')
img = keypoint_detection(frame, detector, net, ctx=ctx)
cv_plot_image(img)
if isinstance(img, mx.nd.NDArray):
img = img.asnumpy()
canvas = cv2.cvtColor(img, cv2.COLOR_RGB2BGR)
result.write(canvas)
else:
break
if cv2.waitKey(1) == 27:
cv2.destroyAllWindows()
break
cap.release()
cv2.destroyAllWindows()
result.release()
def keypoint_detection(img, detector, pose_net, ctx=mx.gpu()):
x, scaled_img = gcv.data.transforms.presets.yolo.transform_test(img, short=480, max_size=1024)
x = x.as_in_context(ctx)
class_IDs, scores, bounding_boxs = detector(x)
pose_input, upscale_bbox = detector_to_alpha_pose(scaled_img, class_IDs, scores, bounding_boxs,ctx=ctx)
if len(upscale_bbox) > 0:
predicted_heatmap = pose_net(pose_input)
pred_coords, confidence = heatmap_to_coord_alpha_pose(predicted_heatmap, upscale_bbox)
scale = 1.0 * img.shape[0] / scaled_img.shape[0]
img = cv_plot_keypoints(img.asnumpy(), pred_coords, confidence, class_IDs, bounding_boxs, scores, box_thresh=0.5, keypoint_thresh=0.2, scale=scale)
return img
If I use the Simple Pose Model it works perfecty.
|
closed
|
2021-09-30T19:45:23Z
|
2021-10-13T22:45:08Z
|
https://github.com/dmlc/gluon-cv/issues/1708
|
[] |
handreyeg
| 2
|
assafelovic/gpt-researcher
|
automation
| 970
|
"Failed to load any documents!"
|
**Describe the bug**
I'm getting the error below after researching something via a local installation of GPTResearcher:
"Failed to load any documents!"
**To Reproduce**
Steps to reproduce the behavior:
1. I install the project following the [Getting Started](https://docs.gptr.dev/docs/gpt-researcher/getting-started/getting-started) guide.
2. Added these keys to my .env file: OPENAI_API_KEY=xxx, TAVILY_API_KEY=xxx, RETRIEVER=tavily
3. Launch the app via http://localhost:8000 and start a research with the default field selected including Source = "The Web".
**Expected behavior**
I should get the research result
**Screenshots**

**Desktop (please complete the following information):**
- OS: Windows 11
- Browser: Google Chrome
- 130.0.6723.70
**Additional context**
Why is it looking for documents if I selected "The Web" in the source dropdown field?
The ./my-docs folder exists and I added a test.txt file to that folder to see if that would make any difference, but I still get the same error.
|
closed
|
2024-10-31T19:12:59Z
|
2025-02-06T19:34:28Z
|
https://github.com/assafelovic/gpt-researcher/issues/970
|
[] |
badcom
| 2
|
amidaware/tacticalrmm
|
django
| 2,161
|
Probleme with connection between Tactical and Mesh Central
|
**Server Info (please complete the following information):**
- OS: Ubuntu 20.04
- Browser: chrome & firefox
- RMM Version (as shown in top left of web UI): V1.0.0
**Installation Method:**
- Standard
**Describe the bug**
Hello, I have an issue with the connection between Tactical and Mesh Central.
User synchronization is enabled. When I log in as a local super user in Mesh, I can see that the accounts are created, for example:
user//jmadmin___3 → in Mesh
jmadmin → in TRMM
I can successfully log into Tactical using the username jmadmin,
but I can't log into Mesh—only the local user can access it.
When I use TRMM with the jmadmin account and take remote control or perform other actions, it correctly uses jmadmin via Mesh.
I don't understand why I can't log directly into Mesh. I have assigned the jmadmin user as a full administrator...
|
closed
|
2025-03-06T11:44:16Z
|
2025-03-07T19:01:36Z
|
https://github.com/amidaware/tacticalrmm/issues/2161
|
[] |
Abygail007
| 2
|
polakowo/vectorbt
|
data-visualization
| 21
|
Backtest strategy with prices and signs
|
I have tried to use the package today but it's hard to find a proper function for the problem.
I have simple set up. I am investing in SPY with a minute resolution. I have already calculate signs using ML methods. Now, I would like to backtest my strategy. So, I have a simple data frame with two columns: prices (or returns) and sings (1 and 0, where 1 is hold and 0 do not hold). Is there some simple function to backtest this kind of strategy. S we have price or return vector and signs vector.
|
closed
|
2020-05-26T08:52:11Z
|
2020-06-01T13:10:06Z
|
https://github.com/polakowo/vectorbt/issues/21
|
[] |
MislavSag
| 7
|
MaartenGr/BERTopic
|
nlp
| 1,760
|
Saving parameters and results to a log file
|
I found useful to save the parameters and results to a log file.
I extended the BERTopic class to fit my needs, here you can have a look:
https://github.com/buscon/fg_analysis_with_BERT/blob/main/classes/custom_log_bertopic.py
It is a barebone implementation, it should be extended and refined.
If you @MaartenGr are interested to integrate such a feature in BERTopic, I can fork it and implement this feature inside the BERTopic structure and make a PR.
|
open
|
2024-01-17T20:22:10Z
|
2024-01-29T03:31:30Z
|
https://github.com/MaartenGr/BERTopic/issues/1760
|
[] |
buscon
| 5
|
aimhubio/aim
|
data-visualization
| 2,501
|
Flag / option to auto-commit or store diff patch
|
## 🚀 Feature
Flag or option on run instantiation (or maybe some config file somewhere) to auto-commit when a new run is started so that commits stored on Aim are synced with the git repo.
### Motivation
Often, commits on Aim are not in sync with the git repo state because uncommitted changes are not incorporated.
### Pitch
Let's auto-commit or store a diff patch on Aim so that these changes are reflected on Aim.
### Alternatives
N/A
### Additional context
N/A
|
open
|
2023-01-25T19:13:09Z
|
2023-02-01T18:47:04Z
|
https://github.com/aimhubio/aim/issues/2501
|
[
"type / enhancement",
"area / SDK-storage"
] |
rodrigo-castellon
| 1
|
proplot-dev/proplot
|
data-visualization
| 271
|
`latinline=True` and `lonlines` lead to infinity
|
### Description
If we set `latinline=True` and `lonlines=xxxxx` at the same time, gridliner will get float infinity.
### Steps to reproduce
```python
import proplot as pplt
fig, ax = pplt.subplots(proj='pcarree')
ax.format(coast=True, lonlabels=True, labels=True, latlines=10, lonlines=20, latinline=True)
```
**Actual behavior**:
```
OverflowError Traceback (most recent call last)
~/new/miniconda3/envs/pyresample_min/lib/python3.8/site-packages/IPython/core/formatters.py in __call__(self, obj)
339 pass
340 else:
--> 341 return printer(obj)
342 # Finally look for special method names
343 method = get_real_method(obj, self.print_method)
~/new/miniconda3/envs/pyresample_min/lib/python3.8/site-packages/IPython/core/pylabtools.py in <lambda>(fig)
253 png_formatter.for_type(Figure, lambda fig: print_figure(fig, 'png', **kwargs))
254 if 'retina' in formats or 'png2x' in formats:
--> 255 png_formatter.for_type(Figure, lambda fig: retina_figure(fig, **kwargs))
256 if 'jpg' in formats or 'jpeg' in formats:
257 jpg_formatter.for_type(Figure, lambda fig: print_figure(fig, 'jpg', **kwargs))
~/new/miniconda3/envs/pyresample_min/lib/python3.8/site-packages/IPython/core/pylabtools.py in retina_figure(fig, **kwargs)
143 def retina_figure(fig, **kwargs):
144 """format a figure as a pixel-doubled (retina) PNG"""
--> 145 pngdata = print_figure(fig, fmt='retina', **kwargs)
146 # Make sure that retina_figure acts just like print_figure and returns
147 # None when the figure is empty.
~/new/miniconda3/envs/pyresample_min/lib/python3.8/site-packages/IPython/core/pylabtools.py in print_figure(fig, fmt, bbox_inches, **kwargs)
135 FigureCanvasBase(fig)
136
--> 137 fig.canvas.print_figure(bytes_io, **kw)
138 data = bytes_io.getvalue()
139 if fmt == 'svg':
~/new/miniconda3/envs/pyresample_min/lib/python3.8/site-packages/proplot/figure.py in _canvas_preprocess(self, *args, **kwargs)
463 ctx3 = rc.context(fig._mathtext_context) # draw with figure-specific setting
464 with ctx1, ctx2, ctx3:
--> 465 fig.auto_layout()
466 return func(self, *args, **kwargs)
467
~/new/miniconda3/envs/pyresample_min/lib/python3.8/site-packages/proplot/figure.py in auto_layout(self, renderer, aspect, tight, resize)
1395 _align_content()
1396 if tight:
-> 1397 gs._auto_layout_space(renderer)
1398 _align_content()
1399
~/new/miniconda3/envs/pyresample_min/lib/python3.8/site-packages/proplot/gridspec.py in _auto_layout_space(self, renderer)
845 pad = self._outerpad
846 obox = fig.bbox_inches # original bbox
--> 847 bbox = fig.get_tightbbox(renderer)
848
849 # Calculate new figure margins
~/new/miniconda3/envs/pyresample_min/lib/python3.8/site-packages/matplotlib/figure.py in get_tightbbox(self, renderer, bbox_extra_artists)
2504
2505 for a in artists:
-> 2506 bbox = a.get_tightbbox(renderer)
2507 if bbox is not None and (bbox.width != 0 or bbox.height != 0):
2508 bb.append(bbox)
~/new/miniconda3/envs/pyresample_min/lib/python3.8/site-packages/proplot/axes/geo.py in get_tightbbox(self, renderer, *args, **kwargs)
1082 for gl in self._gridliners:
1083 if _version_cartopy >= 0.18:
-> 1084 gl._draw_gridliner(renderer=renderer)
1085 else:
1086 gl._draw_gridliner(background_patch=self.background_patch)
~/new/miniconda3/envs/pyresample_min/lib/python3.8/site-packages/proplot/axes/geo.py in _draw_gridliner(self, *args, **kwargs)
760 # the time between v0.17 and v0.18 is any indication.
761 def _draw_gridliner(self, *args, **kwargs):
--> 762 result = type(self)._draw_gridliner(self, *args, **kwargs)
763 if _version_cartopy >= 0.18:
764 lon_lim, _ = self._axes_domain()
~/new/miniconda3/envs/pyresample_min/lib/python3.8/site-packages/cartopy/mpl/gridliner.py in _draw_gridliner(self, nx, ny, renderer)
513 y_midpoints = self._find_midpoints(lat_lim, lat_ticks)
514 if self.y_inline:
--> 515 x_midpoints = self._find_midpoints(lon_lim, lon_ticks)
516
517 for lonlat, lines, line_ticks, formatter, label_style in (
~/new/miniconda3/envs/pyresample_min/lib/python3.8/site-packages/cartopy/mpl/gridliner.py in _find_midpoints(self, lim, ticks)
408 lq = 25
409 uq = 75
--> 410 midpoints = (self._round(np.percentile(lim, lq), cent),
411 self._round(np.percentile(lim, uq), cent))
412 return midpoints
~/new/miniconda3/envs/pyresample_min/lib/python3.8/site-packages/cartopy/mpl/gridliner.py in _round(x, base)
394 if np.isnan(base):
395 base = 5
--> 396 return int(base * round(x / base))
397
398 def _find_midpoints(self, lim, ticks):
OverflowError: cannot convert float infinity to integer
```
### Version
matplotlib 3.3.4
proplot 0.8.1
|
closed
|
2021-08-31T08:01:49Z
|
2021-09-04T00:20:52Z
|
https://github.com/proplot-dev/proplot/issues/271
|
[
"dependencies"
] |
zxdawn
| 1
|
sqlalchemy/sqlalchemy
|
sqlalchemy
| 10,170
|
PyHive, Presto connector returning wrong resultset
|
### Describe the bug
I'm using Presto Cluster for processing large amount of data.
To visualize the data I use the connector provided and suggested by the official Superset documentation, which is PyHive from the SQLAlchemy library and I'm using the default settings for the connection.
When using the provided pyhive presto connector and executing a very simple query - "SELECT * FROM test_table", the returned number of rows by the resultset is incorrect compared with the same query executed in the presto-cli app, the official connector provided by the Presto documentation.
I created two simple python scripts to test Presto connection using PyHive and the official jdbc.jar driver.
The PyHive connector returned wrong number of rows in the resultset about 817000 rows, exactly the same number of rows that was returned by the Superset chart. The connector with the official jdbc driver returned the correct amount of data - 875000 rows.
It looks like the issue is caused by the PyHive connector. Is it possible to change the connection method from PyHive to the official JDBC driver?
I'm attaching the two python scripts that I used to reproduce the issue.
```
#This Python script is using PyHive
from pyhive import presto
def execute_presto_query(host, port, user, catalog, schema, table, max_rows):
connection = presto.connect(host=host, port=port, username=user, catalog=catalog, schema=schema, session_props={'query_max_output_size': '1TB'})
try:
cursor = connection.cursor()
query = f"""SELECT * FROM test_table"""
cursor.execute(query)
total_rows = 0
while True:
rows = cursor.fetchmany(max_rows)
if not rows:
break
for row in rows:
total_rows += 1
print(row)
except Exception as e:
print("Error executing the query:", e)
finally:
print(total_rows)
cursor.close()
connection.close()
if __name__ == "__main__":
host = "localhost"
port = 30000
user = "testUser"
catalog = "pinot"
schema = "default"
table = "test_table"
max_rows = 1000000
execute_presto_query(host, port, user, catalog, schema, table, max_rows)
```
```
#This Python script is using the official JDBC driver
import jaydebeapi
import jpype
def execute_presto_query(host, port, user, catalog, schema, table, max_rows):
jar_file = '/home/admin1/Downloads/presto-jdbc-0.282.jar'
jpype.startJVM(jpype.getDefaultJVMPath(), "-Djava.class.path=" + jar_file)
connection_url = f'jdbc:presto://{host}:{port}/{catalog}/{schema}'
conn = jaydebeapi.connect(
'com.facebook.presto.jdbc.PrestoDriver',
connection_url,
{'user': user},
jar_file
)
try:
cursor = conn.cursor()
query = f"SELECT * FROM test_table"
cursor.execute(query)
rows = cursor.fetchall()
for row in rows:
print(row)
print(f"Total rows returned: {len(rows)}")
except Exception as e:
print("Error executing the query:", e)
finally:
cursor.close()
conn.close()
jpype.shutdownJVM()
if __name__ == "__main__":
host = "localhost"
port = 30000
user = "testUsername"
catalog = "pinot"
schema = "default"
table = "test_table"
max_rows = 1000000
execute_presto_query(host, port, user, catalog, schema, table, max_rows)
```
### Optional link from https://docs.sqlalchemy.org which documents the behavior that is expected
_No response_
### SQLAlchemy Version in Use
Not sure which version is used in the latest stable release of Superset
### DBAPI (i.e. the database driver)
PyHive
### Database Vendor and Major Version
Presto Cluster connected to Apache Pinot
### Python Version
3.8.13
### Operating system
Linux
### To Reproduce
```python
The code was provided in the description of the issue.
```
### Error
No error was returned by the Presto Cluster.
### Additional context
_No response_
|
closed
|
2023-08-01T07:41:22Z
|
2023-08-01T12:05:54Z
|
https://github.com/sqlalchemy/sqlalchemy/issues/10170
|
[
"external driver issues"
] |
alextk87
| 3
|
iMerica/dj-rest-auth
|
rest-api
| 564
|
Custom Register serializer not detected after switching from django-rest-auth
|
Hi !
I'm upgrading a project from Django 3.2.9 to 4.2.5, in a docker container.
Previous project `requirements.txt` :
```
Django==3.2.9
djangorestframework==3.12.4
django-allauth==0.46.0
django-rest_auth==0.9.5
```
I upgraded to
```
Django==4.2.5
djangorestframework==3.14.0
django-allauth==0.58.1
dj-rest-auth==5.0.1
```
I switched to `dj-rest-auth` as asked in `django-rest-auth` https://github.com/Omaraitbenhaddi/django-rest-auth
I only changed the imports and the name usage of the packages and run a `migrate` command.
Expected behavior: Project keep using `CustomRegisterSerializer` as before
Actual behavior: Now, project ignore `CustomRegisterSerializer` and uses regular `RegisterSerializer` instead.
All resources online point to the `settings,py` but I have the correct `REST_AUTH_REGISTER_SERIALIZERS` (I even tried to put my serializer in `REST_AUTH_SERIALIZERS` instead).
I also uninstalled and reinstalled `allauth`. But keep getting the same issue:
```
File "/app/cloud_api/apps/user/adapters.py", line 13, in save_user
user.first_name = data['first_name']
KeyError: 'first_name'
```
Because wrong Serializer is used. Any idea of what I did wrong in the migration ?
`serializers.py`
```
from django.contrib.auth import get_user_model
from django.core.exceptions import ObjectDoesNotExist
from django.utils.translation import gettext_lazy as _
from rest_framework import serializers
from dry_rest_permissions.generics import DRYGlobalPermissionsField
from dj_rest_auth.registration.serializers import RegisterSerializer
from dj_rest_auth.serializers import PasswordResetSerializer
User = get_user_model()
class CustomRegisterSerializer(RegisterSerializer):
first_name = serializers.CharField(
max_length=30,
allow_blank=True,
allow_null=True,
required=False
)
last_name = serializers.CharField(
max_length=150,
allow_blank=True,
allow_null=True,
required=False
)
password1 = serializers.CharField(write_only=True, required=False)
password2 = serializers.CharField(write_only=True, required=False)
def get_cleaned_data(self):
return {
'username': self.validated_data.get('username', ''),
'password1': self.validated_data.get('password1', ''),
'email': self.validated_data.get('email', ''),
'first_name': self.validated_data.get('first_name', ''),
'last_name': self.validated_data.get('last_name', ''),
}
def validate(self, data):
if 'password1' in data and data['password1'] != data['password2']:
raise serializers.ValidationError(
_("The two password didn't match."))
return data
```
`adapaters.py`
```
from allauth.account.adapter import DefaultAccountAdapter
from django.contrib.auth import get_user_model
User = get_user_model()
class AccountAdapter(DefaultAccountAdapter):
def save_user(self, request, user, form, commit=True):
data = form.cleaned_data
user.username = data['username']
user.email = data['email']
user.first_name = data['first_name']
user.last_name = data['last_name']
if 'password1' in data:
user.set_password(data['password1'])
else:
user.set_unusable_password()
self.populate_username(request, user)
if commit:
user.save()
return user
```
`urls.py`
```
urlpatterns = [
path(
'rest-auth/',
include('dj_rest_auth.urls')
),
path(
"password-reset/confirm/<uidb64>/<token>/",
TemplateView.as_view(template_name="password_reset_confirm.html"),
name='password_reset_confirm'
),
re_path(r'^accounts/', include('allauth.urls'), name='socialaccount_signup'),
path(
'rest-auth/registration/',
include('dj_rest_auth.registration.urls')
),
path('dj-rest-auth/facebook/', FacebookLogin.as_view(), name='fb_login'),
path('', include(router.urls)), # includes router generated URL
]
```
`settings.py`
```
INSTALLED_APPS = [
. . .
'dj_rest_auth',
'allauth',
'allauth.account',
'dj_rest_auth.registration',
'allauth.socialaccount',
'allauth.socialaccount.providers.facebook',
. . .
]
MIDDLEWARE = [
. . .
'allauth.account.middleware.AccountMiddleware',
]
ACCOUNT_ADAPTER = 'cloud_api'\
'.apps.user.adapters.AccountAdapter'
REST_AUTH_SERIALIZERS = {
'USER_DETAILS_SERIALIZER':
'cloud_api'
'.apps.user.serializers.UserSerializer',
'PASSWORD_RESET_SERIALIZER':
'cloud_api'
'.apps.user.serializers.CustomPasswordResetSerializer',
}
REST_AUTH_REGISTER_SERIALIZERS = {
'REGISTER_SERIALIZER':
'cloud_api'
'.apps.user.serializers.CustomRegisterSerializer'
}
```
|
closed
|
2023-11-02T14:07:35Z
|
2023-11-02T14:47:51Z
|
https://github.com/iMerica/dj-rest-auth/issues/564
|
[] |
RomainFayolle
| 1
|
encode/uvicorn
|
asyncio
| 1,501
|
Gracefully handle HTTP/2 upgrade request
|
I know that HTTP/2 is out of scope of this project. But I am not asking to support HTTP/2.
More and more http clients try to upgrade the connection to HTTP/2. Uvicorn is free to not honor this request. Unfortunately, instead of ignoring the upgrade request, Uvicorn responds with status `400 Bad Request` and the message "Unsupported upgrade request".
According to https://developer.mozilla.org/en-US/docs/Web/HTTP/Protocol_upgrade_mechanism the server should just ignore the upgrade request:
> If the server decides to upgrade the connection, it sends back a [101 Switching Protocols](https://developer.mozilla.org/en-US/docs/Web/HTTP/Status/101) response status with an Upgrade header that specifies the protocol(s) being switched to. If it does not (or cannot) upgrade the connection, it ignores the Upgrade header and sends back a regular response (for example, a [200 OK](https://developer.mozilla.org/en-US/docs/Web/HTTP/Status/200)).
We continue to encounter this issue because the `HttpClient` class of modern OpenJDK versions tries to upgrade the connection to HTTP/2 by default. Uvicorn should just ignore these headers and process the requests as if these headers were not present.
|
closed
|
2022-05-25T12:36:51Z
|
2022-10-19T06:47:31Z
|
https://github.com/encode/uvicorn/issues/1501
|
[
"bug",
"help wanted",
"http"
] |
ChristianCiach
| 7
|
python-arq/arq
|
asyncio
| 469
|
How to monitor
|
open
|
2024-07-11T01:17:25Z
|
2024-07-11T01:17:25Z
|
https://github.com/python-arq/arq/issues/469
|
[] |
yuanjie-ai
| 0
|
|
Guovin/iptv-api
|
api
| 597
|
docker下运行出现不支持IPV6的提示
|
是不是要在docker项目内设置ipv6的内网地址?
目前项目的默认网络模式是桥接模式,是否要改成host模式?尝试改host会出现8000端口被占用。
电脑运行exe客户端会出现弹窗问题
|
closed
|
2024-11-29T14:46:50Z
|
2024-12-02T11:23:03Z
|
https://github.com/Guovin/iptv-api/issues/597
|
[
"duplicate"
] |
FRANKASEE
| 6
|
onnx/onnx
|
pytorch
| 6,014
|
Replace const string references with string_views
|
https://en.cppreference.com/w/cpp/string/basic_string_view string_view is the new recommended way to a function to accept a read-only view of a string. If helps reduce and unify overloads for `const char*` and `const std::string&`, and improves readability.
cc @gramalingam @BowenBao @edgchen1 for comments.
|
closed
|
2024-03-11T18:46:52Z
|
2024-03-15T16:04:08Z
|
https://github.com/onnx/onnx/issues/6014
|
[
"topic: enhancement",
"topic: better engineering"
] |
justinchuby
| 1
|
holoviz/panel
|
matplotlib
| 6,854
|
Landuse classification example error
|
I am trying to reproduce the example at
https://examples.holoviz.org/gallery/landuse_classification/Image_Classification.html
#### ALL software version info
bokeh=3.4.1=pyhd8ed1ab_0
holoviews=1.18.3=pyhd8ed1ab_0
hvplot=0.10.0=pyhd8ed1ab_0
intake=2.0.5=pyhd8ed1ab_0
intake-xarray=0.7.0=pyhd8ed1ab_0
jupyterlab=4.1.8=pyhd8ed1ab_0
pandas=2.2.2=py39haf03413_0
panel=1.4.2=pyhd8ed1ab_0
python=3.9.19=h7a9c478_0_cpython
rasterio=1.3.9=py39h69ae74f_2
rioxarray=0.15.0=pyhd8ed1ab_0
xarray=2024.3.0=pyhd8ed1ab_0
#### Description of expected behavior and the observed behavior
According to the example, once the S3 AWS intake catalog is open, users should gain access to an xarray.DataArray. Instead, a FileNotFoundError occurs.
#### Complete, minimal, self-contained example code that reproduces the issue
```
# code goes here between backticks
cat = intake.open_catalog('https://s3.amazonaws.com/earth-data/UCMerced_LandUse/catalog.yml')
da = cat.UCMerced_LandUse_all().to_dask()
```
#### Stack traceback and/or browser JavaScript console output
FileNotFoundError: earth-data[/UCMerced_LandUse/Images/](http://localhost:8889/UCMerced_LandUse/Images/){landuse}[/](http://localhost:8889/){}{id:2d}.tif
|
open
|
2024-05-20T21:05:12Z
|
2024-05-21T21:22:50Z
|
https://github.com/holoviz/panel/issues/6854
|
[] |
ials
| 4
|
slackapi/bolt-python
|
fastapi
| 667
|
Unable to set true in a modal
|
I am having issues using `true` anywhere in the view payload. Instead I am getting the following error in the console when trying to open the view: `NameError: name 'true' is not defined`
### Reproducible in:
```python
@app.shortcut("request")
def open_modal(ack, shortcut, client):
ack()
client.views_open(
trigger_id=shortcut["trigger_id"],
# A simple view payload for a modal
view={
"type": "modal",
"callback_id": "request_view",
"title": {"type": "plain_text", "text": "Submit Request"},
"close": {"type": "plain_text", "text": "Close"},
"submit": {"type": "plain_text", "text": "Submit"},
"blocks": [
{
"type": "input",
"element": {
"type": "plain_text_input",
"action_id": "subject"
},
"label": {
"type": "plain_text",
"text": "Subject",
"emoji": true
}
},
]
}
)
```
#### The `slack_bolt` version
(Paste the output of `pip freeze | grep slack`)
1.14.0
#### Python runtime version
(Paste the output of `python --version`)
Python 3.9.10
#### OS info
ProductName: macOS
ProductVersion: 11.5.2
BuildVersion: 20G95
Darwin Kernel Version 20.6.0: Wed Jun 23 00:26:31 PDT 2021
#### Steps to reproduce:
1. Run example and try to open the shortcut.
### Expected result:
The input should have the ability to add emojis in text input.
### Actual result:
```
Error in console:
2022-06-05 20:20:12,499 Failed to run listener function (error: name 'true' is not defined)
Traceback (most recent call last):
File "/Users/user/Dev/myslackapp/.venv/lib/python3.9/site-packages/slack_bolt/listener/thread_runner.py", line 65, in run
returned_value = listener.run_ack_function(
File "/Users/user/Dev/myslackapp/.venv/lib/python3.9/site-packages/slack_bolt/listener/custom_listener.py", line 50, in run_ack_function
return self.ack_function(
File "/Users/user/Dev/myslackapp/./handler.py", line 45, in open_modal
"emoji": true
NameError: name 'true' is not defined
```
## Requirements
Please read the [Contributing guidelines](https://github.com/slackapi/bolt-python/blob/main/.github/contributing.md) and [Code of Conduct](https://slackhq.github.io/code-of-conduct) before creating this issue or pull request. By submitting, you are agreeing to those rules.
|
closed
|
2022-06-06T00:29:36Z
|
2022-06-06T21:28:14Z
|
https://github.com/slackapi/bolt-python/issues/667
|
[
"question"
] |
BMonsalvatge
| 2
|
keras-team/keras
|
machine-learning
| 20,833
|
Keras 2.15 is unable to load "h5" dumps created by itself (but can load models made in 2.12)
|
Using keras 2.15 installed with tensorflow 2.15, I'm taking a sample code from keras documentation: https://keras.io/guides/serialization_and_saving/ with the only change - I'm saving "h5" file instead of "keras".
Sample code produces output:
```
numpy: 1.26.4
tensorflow: 2.15.1
keras: 2.15.0
TypeError: Error when deserializing class 'Dense' using config={'name': 'dense', 'trainable': True, 'dtype': 'float32', 'units': 1, 'activation': {'module': 'builtins', 'class_name': 'function', 'config': 'my_package>custom_fn', 'registered_name': 'function'}, 'use_bias': True, 'kernel_initializer': {'module': 'keras.initializers', 'class_name': 'GlorotUniform', 'config': {'seed': None}, 'registered_name': None}, 'bias_initializer': {'module': 'keras.initializers', 'class_name': 'Zeros', 'config': {}, 'registered_name': None}, 'kernel_regularizer': None, 'bias_regularizer': None, 'activity_regularizer': None, 'kernel_constraint': None, 'bias_constraint': None}.
Exception encountered: Unknown activation function: 'function'. Please ensure you are using a `keras.utils.custom_object_scope` and that this object is included in the scope. See https://www.tensorflow.org/guide/keras/save_and_serialize#registering_the_custom_object for details.
```
Sample code:
```python
import numpy as np
import tensorflow as tf
import keras
print("numpy:", np.__version__)
print("tensorflow:", tf.__version__)
print("keras:", keras.__version__)
keras.saving.get_custom_objects().clear()
@keras.saving.register_keras_serializable(package="MyLayers")
class CustomLayer(keras.layers.Layer):
def __init__(self, factor):
super().__init__()
self.factor = factor
def call(self, x):
return x * self.factor
def get_config(self):
return {"factor": self.factor}
@keras.saving.register_keras_serializable(package="my_package", name="custom_fn")
def custom_fn(x):
return x**2
# Create the model.
def get_model():
inputs = keras.Input(shape=(4,))
mid = CustomLayer(0.5)(inputs)
outputs = keras.layers.Dense(1, activation=custom_fn)(mid)
model = keras.Model(inputs, outputs)
model.compile(optimizer="rmsprop", loss="mean_squared_error")
return model
# Train the model.
def train_model(model):
input = np.random.random((4, 4))
target = np.random.random((4, 1))
model.fit(input, target)
return model
if __name__ == "__main__":
# This is the only difference wit the documentation
# when using "keras", loading succeeds.
file_format = "h5"
file_name = f"custom_model_reg.{file_format}"
model = get_model()
model = train_model(model)
model.save(file_name)
# Raises error
reconstructed_model = keras.models.load_model(file_name)
```
If I create this model in keras 2.12, loading succeeds.
Comparing metadata for this model, created in 2.12 and 2.15, there is a certain difference:
Here is 2.12 metadata:
```json
{
"class_name": "Dense",
"config": {
"name": "dense",
"trainable": true,
"dtype": "float32",
"units": 1,
"activation": "custom_fn",
...
```
and here is 2.15:
```json
"class_name": "Dense",
"config": {
"name": "dense",
"trainable": true,
"dtype": "float32",
"units": 1,
"activation": {
"module": "builtins",
"class_name": "function",
"config": "custom_fn",
"registered_name": "function"
},
...
```
2.15 changed "activation" definition from string to dictionary.
Further debugging shows that when we try to load "h5" file, execution eventually reaches function `keras.src.saving.legacy.serialization.class_and_config_for_serialized_keras_object`, which takes only "class_name" to resolve the object, and, naturally, fails, because class_name is "function":
```python
class_name = config["class_name"]
cls = object_registration.get_registered_object(
class_name, custom_objects, module_objects
)
if cls is None:
raise ValueError(
f"Unknown {printable_module_name}: '{class_name}'. "
```
So the question is - is there a way to fix this or at least workaround?
tensorflow 2.15 is highest version available to me.
|
closed
|
2025-01-31T12:51:41Z
|
2025-03-06T02:04:46Z
|
https://github.com/keras-team/keras/issues/20833
|
[
"stat:awaiting response from contributor",
"stale",
"type:Bug"
] |
nchaly
| 3
|
axnsan12/drf-yasg
|
django
| 124
|
I have a little update about the summary
|
32, 43, 336...341 rows. Thank you
```from collections import OrderedDict
from rest_framework.request import is_form_media_type
from rest_framework.schemas import AutoSchema
from rest_framework.status import is_success
from .. import openapi
from ..errors import SwaggerGenerationError
from ..utils import (
force_serializer_instance, get_consumes, get_produces, guess_response_status, is_list_view, no_body,
param_list_to_odict
)
from .base import ViewInspector
class SwaggerAutoSchema(ViewInspector):
def __init__(self, view, path, method, components, request, overrides):
super(SwaggerAutoSchema, self).__init__(view, path, method, components, request, overrides)
self._sch = AutoSchema()
self._sch.view = view
def get_operation(self, operation_keys):
consumes = self.get_consumes()
produces = self.get_produces()
body = self.get_request_body_parameters(consumes)
query = self.get_query_parameters()
parameters = body + query
parameters = [param for param in parameters if param is not None]
parameters = self.add_manual_parameters(parameters)
summary = self.get_operation_id()
operation_id = self.get_operation_id(operation_keys)
description = self.get_description()
security = self.get_security()
assert security is None or isinstance(security, list), "security must be a list of securiy requirement objects"
tags = self.get_tags(operation_keys)
responses = self.get_responses()
return openapi.Operation(
summary=summary,
operation_id=operation_id,
description=description,
responses=responses,
parameters=parameters,
consumes=consumes,
produces=produces,
tags=tags,
security=security
)
def get_request_body_parameters(self, consumes):
"""Return the request body parameters for this view. |br|
This is either:
- a list with a single object Parameter with a :class:`.Schema` derived from the request serializer
- a list of primitive Parameters parsed as form data
:param list[str] consumes: a list of accepted MIME types as returned by :meth:`.get_consumes`
:return: a (potentially empty) list of :class:`.Parameter`\ s either ``in: body`` or ``in: formData``
:rtype: list[openapi.Parameter]
"""
serializer = self.get_request_serializer()
schema = None
if serializer is None:
return []
if isinstance(serializer, openapi.Schema.OR_REF):
schema = serializer
if any(is_form_media_type(encoding) for encoding in consumes):
if schema is not None:
raise SwaggerGenerationError("form request body cannot be a Schema")
return self.get_request_form_parameters(serializer)
else:
if schema is None:
schema = self.get_request_body_schema(serializer)
return [self.make_body_parameter(schema)] if schema is not None else []
def get_view_serializer(self):
"""Return the serializer as defined by the view's ``get_serializer()`` method.
:return: the view's ``Serializer``
"""
if not hasattr(self.view, 'get_serializer'):
return None
return self.view.get_serializer()
def get_request_serializer(self):
"""Return the request serializer (used for parsing the request payload) for this endpoint.
:return: the request serializer, or one of :class:`.Schema`, :class:`.SchemaRef`, ``None``
"""
body_override = self.overrides.get('request_body', None)
if body_override is not None:
if body_override is no_body:
return None
if self.method not in self.body_methods:
raise SwaggerGenerationError("request_body can only be applied to (" + ','.join(self.body_methods) +
"); are you looking for query_serializer or manual_parameters?")
if isinstance(body_override, openapi.Schema.OR_REF):
return body_override
return force_serializer_instance(body_override)
elif self.method in self.implicit_body_methods:
return self.get_view_serializer()
return None
def get_request_form_parameters(self, serializer):
"""Given a Serializer, return a list of ``in: formData`` :class:`.Parameter`\ s.
:param serializer: the view's request serializer as returned by :meth:`.get_request_serializer`
:rtype: list[openapi.Parameter]
"""
return self.serializer_to_parameters(serializer, in_=openapi.IN_FORM)
def get_request_body_schema(self, serializer):
"""Return the :class:`.Schema` for a given request's body data. Only applies to PUT, PATCH and POST requests.
:param serializer: the view's request serializer as returned by :meth:`.get_request_serializer`
:rtype: openapi.Schema
"""
return self.serializer_to_schema(serializer)
def make_body_parameter(self, schema):
"""Given a :class:`.Schema` object, create an ``in: body`` :class:`.Parameter`.
:param openapi.Schema schema: the request body schema
:rtype: openapi.Parameter
"""
return openapi.Parameter(name='data', in_=openapi.IN_BODY, required=True, schema=schema)
def add_manual_parameters(self, parameters):
"""Add/replace parameters from the given list of automatically generated request parameters.
:param list[openapi.Parameter] parameters: genereated parameters
:return: modified parameters
:rtype: list[openapi.Parameter]
"""
parameters = param_list_to_odict(parameters)
manual_parameters = self.overrides.get('manual_parameters', None) or []
if any(param.in_ == openapi.IN_BODY for param in manual_parameters): # pragma: no cover
raise SwaggerGenerationError("specify the body parameter as a Schema or Serializer in request_body")
if any(param.in_ == openapi.IN_FORM for param in manual_parameters): # pragma: no cover
if any(param.in_ == openapi.IN_BODY for param in parameters.values()):
raise SwaggerGenerationError("cannot add form parameters when the request has a request body; "
"did you forget to set an appropriate parser class on the view?")
if self.method not in self.body_methods:
raise SwaggerGenerationError("form parameters can only be applied to (" + ','.join(self.body_methods) +
") HTTP methods")
parameters.update(param_list_to_odict(manual_parameters))
return list(parameters.values())
def get_responses(self):
"""Get the possible responses for this view as a swagger :class:`.Responses` object.
:return: the documented responses
:rtype: openapi.Responses
"""
response_serializers = self.get_response_serializers()
return openapi.Responses(
responses=self.get_response_schemas(response_serializers)
)
def get_default_responses(self):
"""Get the default responses determined for this view from the request serializer and request method.
:type: dict[str, openapi.Schema]
"""
method = self.method.lower()
default_status = guess_response_status(method)
default_schema = ''
if method in ('get', 'post', 'put', 'patch'):
default_schema = self.get_request_serializer() or self.get_view_serializer()
default_schema = default_schema or ''
if any(is_form_media_type(encoding) for encoding in self.get_consumes()):
default_schema = ''
if default_schema and not isinstance(default_schema, openapi.Schema):
default_schema = self.serializer_to_schema(default_schema) or ''
if default_schema:
if is_list_view(self.path, self.method, self.view) and self.method.lower() == 'get':
default_schema = openapi.Schema(type=openapi.TYPE_ARRAY, items=default_schema)
if self.should_page():
default_schema = self.get_paginated_response(default_schema) or default_schema
return OrderedDict({str(default_status): default_schema})
def get_response_serializers(self):
"""Return the response codes that this view is expected to return, and the serializer for each response body.
The return value should be a dict where the keys are possible status codes, and values are either strings,
``Serializer``\ s, :class:`.Schema`, :class:`.SchemaRef` or :class:`.Response` objects. See
:func:`@swagger_auto_schema <.swagger_auto_schema>` for more details.
:return: the response serializers
:rtype: dict
"""
manual_responses = self.overrides.get('responses', None) or {}
manual_responses = OrderedDict((str(sc), resp) for sc, resp in manual_responses.items())
responses = OrderedDict()
if not any(is_success(int(sc)) for sc in manual_responses if sc != 'default'):
responses = self.get_default_responses()
responses.update((str(sc), resp) for sc, resp in manual_responses.items())
return responses
def get_response_schemas(self, response_serializers):
"""Return the :class:`.openapi.Response` objects calculated for this view.
:param dict response_serializers: response serializers as returned by :meth:`.get_response_serializers`
:return: a dictionary of status code to :class:`.Response` object
:rtype: dict[str, openapi.Response]
"""
responses = OrderedDict()
for sc, serializer in response_serializers.items():
if isinstance(serializer, str):
response = openapi.Response(
description=serializer
)
elif isinstance(serializer, openapi.Response):
response = serializer
if hasattr(response, 'schema') and not isinstance(response.schema, openapi.Schema.OR_REF):
serializer = force_serializer_instance(response.schema)
response.schema = self.serializer_to_schema(serializer)
elif isinstance(serializer, openapi.Schema.OR_REF):
response = openapi.Response(
description='',
schema=serializer,
)
else:
serializer = force_serializer_instance(serializer)
response = openapi.Response(
description='',
schema=self.serializer_to_schema(serializer),
)
responses[str(sc)] = response
return responses
def get_query_serializer(self):
"""Return the query serializer (used for parsing query parameters) for this endpoint.
:return: the query serializer, or ``None``
"""
query_serializer = self.overrides.get('query_serializer', None)
if query_serializer is not None:
query_serializer = force_serializer_instance(query_serializer)
return query_serializer
def get_query_parameters(self):
"""Return the query parameters accepted by this view.
:rtype: list[openapi.Parameter]
"""
natural_parameters = self.get_filter_parameters() + self.get_pagination_parameters()
query_serializer = self.get_query_serializer()
serializer_parameters = []
if query_serializer is not None:
serializer_parameters = self.serializer_to_parameters(query_serializer, in_=openapi.IN_QUERY)
if len(set(param_list_to_odict(natural_parameters)) & set(param_list_to_odict(serializer_parameters))) != 0:
raise SwaggerGenerationError(
"your query_serializer contains fields that conflict with the "
"filter_backend or paginator_class on the view - %s %s" % (self.method, self.path)
)
return natural_parameters + serializer_parameters
def get_operation_id(self, operation_keys):
"""Return an unique ID for this operation. The ID must be unique across
all :class:`.Operation` objects in the API.
:param tuple[str] operation_keys: an array of keys derived from the pathdescribing the hierarchical layout
of this view in the API; e.g. ``('snippets', 'list')``, ``('snippets', 'retrieve')``, etc.
:rtype: str
"""
operation_id = self.overrides.get('operation_id', '')
if not operation_id:
operation_id = '_'.join(operation_keys)
return operation_id
def get_description(self):
"""Return an operation description determined as appropriate from the view's method and class docstrings.
:return: the operation description
:rtype: str
"""
description = self.overrides.get('operation_description', None)
if description is None:
description = self._sch.get_description(self.path, self.method)
return description
def get_security(self):
"""Return a list of security requirements for this operation.
Returning an empty list marks the endpoint as unauthenticated (i.e. removes all accepted
authentication schemes). Returning ``None`` will inherit the top-level secuirty requirements.
:return: security requirements
:rtype: list[dict[str,list[str]]]"""
return self.overrides.get('security', None)
def get_tags(self, operation_keys):
"""Get a list of tags for this operation. Tags determine how operations relate with each other, and in the UI
each tag will show as a group containing the operations that use it.
:param tuple[str] operation_keys: an array of keys derived from the pathdescribing the hierarchical layout
of this view in the API; e.g. ``('snippets', 'list')``, ``('snippets', 'retrieve')``, etc.
:rtype: list[str]
"""
return [operation_keys[0]]
def get_consumes(self):
"""Return the MIME types this endpoint can consume.
:rtype: list[str]
"""
return get_consumes(getattr(self.view, 'parser_classes', []))
def get_produces(self):
"""Return the MIME types this endpoint can produce.
:rtype: list[str]
"""
return get_produces(getattr(self.view, 'renderer_classes', []))
def get_summary(self):
"""Return string summary, intended to apply to all operations in this
path. Should be < 120 characters
"""
summary = self.overrides.get('summary', None)
return summary
```
|
closed
|
2018-05-16T11:01:51Z
|
2018-08-06T13:55:43Z
|
https://github.com/axnsan12/drf-yasg/issues/124
|
[] |
Sadygan
| 1
|
statsmodels/statsmodels
|
data-science
| 9,284
|
Should it not be `positive`?
|
Should it not be `positive`?
https://github.com/statsmodels/statsmodels/blob/c2194ff6052362ffc09da7d00a5a9e99ecb11205/statsmodels/tsa/statespace/dynamic_factor_mq.py#L2828
|
closed
|
2024-06-17T04:16:12Z
|
2024-06-26T15:26:26Z
|
https://github.com/statsmodels/statsmodels/issues/9284
|
[] |
Anselmoo
| 0
|
django-oscar/django-oscar
|
django
| 3,874
|
Use `--fail-on-template-vars` to detect broken templates
|
### Issue Summary
There are multiple templates with broken variables, that go unnoticed. E.g.:
- [In the anon order page](https://github.com/django-oscar/django-oscar/blob/master/src/oscar/templates/oscar/customer/anon_order.html#L70), it should be `line.product` instead of `product`
- [In the GA tracking template](https://github.com/django-oscar/django-oscar/blob/master/src/oscar/templates/oscar/partials/google_analytics_transaction.html#L17) order `line.category` is used even though lines don't have a category attribute (anymore)
- [In the thank you page](https://github.com/django-oscar/django-oscar/blob/master/src/oscar/templates/oscar/checkout/thank_you.html#L132) it should be `line.product` instead of `product` as well
- In [base.html](https://github.com/django-oscar/django-oscar/blob/master/src/oscar/templates/oscar/base.html#L4) `LANGUAGE_CODE` is used even though it doesn't exist. that should probably be `request.LANGUAGE_CODE`
- there are probably more
### Current Situation
On small isolated changes like [my recent PR to use the `as_stars` templatetag](https://github.com/django-oscar/django-oscar/pull/3871), code reviews were able to catch these sort of mistakes. I think it was easy to miss the typo or the missing import in the review process though (at least for me 😅)
### Proposed Solution
I believe a better solution would be to use [pytest-django's `--fail-on-template-vars`](https://pytest-django.readthedocs.io/en/latest/usage.html#fail-on-template-vars-fail-for-invalid-variables-in-templates) config to catch these mistakes programmatically.
The biggest "downside" to this feature is that it ignores the `default` templatetag which is used extensively atm. I don't think that's really a problem, it's just a bit more work. On the flipside, it requires you to be very explicit which can be a good thing IMO.
I'd be happy to contribute a PR for this, but would like to get feedback on this proposal beforehand 🙂
|
open
|
2022-02-25T08:31:19Z
|
2023-06-23T14:36:22Z
|
https://github.com/django-oscar/django-oscar/issues/3874
|
[] |
SaturnFromTitan
| 2
|
serengil/deepface
|
machine-learning
| 489
|
Run deepface on GPU
|
Hello
I wonder how we can run the DeepFace library on GPU. I tried your suggestion on other posts, but none of them worked.
I installed TensorFlow-gpu before the DeepFace and set CUDA_VISIBLE_DEVICES on my code. But none of the tricks works. I also tried to install DeepFace with no dependencies but no luck.
When running on Google Colab, the library uses the GPU, but when I tried in my system, it did not.
Can you please advise how to resolve it?
import time
from deepface import DeepFace
from retinaface import RetinaFace
import os
#os.environ["CUDA_DEVICE_ORDER"] = "PCI_BUS_ID"
#os.environ["CUDA_VISIBLE_DEVICES"] = "0,1,2,3"
start = time.time()
faces = RetinaFace.extract_faces("friends2.jpg")
print("There are ", len(faces), " faces in the image")
for face in faces:
obj = DeepFace.analyze(face, detector_backend='skip')
end = time.time()
print("Finished at: ", end - start)
It took 12 second on my machine, but 3 sec on Google Colab.
Python: 3.6.9
tensorflow-gpu: 2.6.2
Ubuntu: 18.04.6 LTS
Thanks
Nadia
|
closed
|
2022-05-27T08:41:56Z
|
2022-05-27T11:33:32Z
|
https://github.com/serengil/deepface/issues/489
|
[
"dependencies"
] |
nadia-maarfavi
| 1
|
encode/apistar
|
api
| 380
|
fields in object type are not required by default as docs says
|
The README says
> Note that child properties are considered to be required if they do not have a default value.
However, check this:
```python
In [3]: class Schema(typesystem.Object):
...: properties = {
...: 'attr1': typesystem.string(max_length=100),
...: 'attr2': typesystem.string(max_length=100),
...: }
...:
...:
In [4]: Schema({'attr1': 'foo'})
Out[4]: {'attr1': 'foo'}
```
As `attr2` has no default, I would expect a validation exception.
I've checked [the code](https://github.com/encode/apistar/blob/7d7dc3a11983915882687ca8607b9eca2ae5ff57/apistar/typesystem.py#L180-L182) and figured that this is what expected
```python
In [5]: class Schema(typesystem.Object):
...: properties = {
...: 'attr1': typesystem.string(max_length=100),
...: 'attr2': typesystem.string(max_length=100),
...: }
...: required = ['attr1', 'attr2']
...:
...:
...:
In [6]: Schema({'attr1': 'foo'})
---------------------------------------------------------------------------
TypeSystemError Traceback (most recent call last)
<ipython-input-6-7426c73bc31e> in <module>()
----> 1 Schema({'attr1': 'foo'})
~/lab/apistar/apistar/typesystem.py in __init__(self, *args, **kwargs)
192
193 if errors:
--> 194 raise TypeSystemError(errors)
195
196
TypeSystemError: {'attr2': 'This field is required.'}
```
so, the implementation is wrong or the docs are outdated?
|
closed
|
2017-12-27T13:00:26Z
|
2018-03-19T13:53:01Z
|
https://github.com/encode/apistar/issues/380
|
[] |
mgaitan
| 1
|
xlwings/xlwings
|
automation
| 2,273
|
pre-commit run --all-files: pip-shims<=0.3.4' does not match '^[a-zA-Z-_.0-9]+$
|
#### OS (e.g. Windows 10 or macOS Sierra)
macOS Monterey
#### Versions of xlwings, Excel and Python (e.g. 0.11.8, Office 365, Python 3.7)
xlwings - 0.30.8 from source
Python 3.9 and 3.11
I did a `pip install -e ".[all]"
seem my `pip list`
```Python
Package Version Editable project location
------------------ -------- -------------------------
appscript 1.2.2
black 23.3.0
blinker 1.6.2
certifi 2023.5.7
cfgv 3.3.1
charset-normalizer 3.1.0
click 8.1.3
contourpy 1.0.7
cycler 0.11.0
distlib 0.3.6
filelock 3.12.0
Flask 2.3.2
fonttools 4.39.4
identify 2.5.24
idna 3.4
iniconfig 2.0.0
isort 5.12.0
itsdangerous 2.1.2
Jinja2 3.1.2
kiwisolver 1.4.4
lxml 4.9.2
MarkupSafe 2.1.2
matplotlib 3.7.1
mypy-extensions 1.0.0
nodeenv 1.8.0
numpy 1.24.3
packaging 23.1
pandas 2.0.2
pathspec 0.11.1
pdfrw 0.4
Pillow 9.5.0
pip 23.1.2
platformdirs 3.5.1
plotly 5.14.1
pluggy 1.0.0
pre-commit 3.3.2
psutil 5.9.5
pyparsing 3.0.9
pytest 7.3.1
python-dateutil 2.8.2
pytz 2023.3
PyYAML 6.0
requests 2.31.0
setuptools 67.7.2
six 1.16.0
tenacity 8.2.2
tzdata 2023.3
urllib3 2.0.2
virtualenv 20.23.0
Werkzeug 2.3.4
wheel 0.40.0
xlwings 0.0.0 /Users/hayer/xlwings
```
#### Describe your issue (incl. Traceback!)
Running `pre-commit` results in some error. It mentions Poetry, although that is not installed. I am not familiar with pre-commit but perhaps is is a issue related to pre-commit.
```python
pre-commit run --all-files
```
#### Include a minimal code sample to reproduce the issue (and attach a sample workbook if required!)
```python
### version information
pre-commit version: 3.3.2
git --version: git version 2.40.1
sys.version:
3.11.3 (main, Apr 7 2023, 20:13:31) [Clang 14.0.0 (clang-1400.0.29.202)]
sys.executable: /Users/hayer/.virtualenvs/xlwings_311/bin/python
os.name: posix
sys.platform: darwin
### error information
An unexpected error has occurred: CalledProcessError: command: ('/Users/hayer/.cache/pre-commit/repojetja5sx/py_env-python3/bin/python', '-mpip', 'install', '.')
return code: 1
stdout:
Processing /Users/hayer/.cache/pre-commit/repojetja5sx
Installing build dependencies: started
Installing build dependencies: finished with status 'done'
Getting requirements to build wheel: started
Getting requirements to build wheel: finished with status 'done'
Preparing metadata (pyproject.toml): started
Preparing metadata (pyproject.toml): finished with status 'error'
stderr:
error: subprocess-exited-with-error
× Preparing metadata (pyproject.toml) did not run successfully.
│ exit code: 1
╰─> [17 lines of output]
Traceback (most recent call last):
File "/Users/hayer/.cache/pre-commit/repojetja5sx/py_env-python3/lib/python3.11/site-packages/pip/_vendor/pyproject_hooks/_in_process/_in_process.py", line 353, in <module>
main()
File "/Users/hayer/.cache/pre-commit/repojetja5sx/py_env-python3/lib/python3.11/site-packages/pip/_vendor/pyproject_hooks/_in_process/_in_process.py", line 335, in main
json_out['return_val'] = hook(**hook_input['kwargs'])
^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/Users/hayer/.cache/pre-commit/repojetja5sx/py_env-python3/lib/python3.11/site-packages/pip/_vendor/pyproject_hooks/_in_process/_in_process.py", line 149, in prepare_metadata_for_build_wheel
return hook(metadata_directory, config_settings)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/private/var/folders/st/_0g2w6450zs5w9xts7h111f00000gn/T/pip-build-env-gq1yq4xp/overlay/lib/python3.11/site-packages/poetry/core/masonry/api.py", line 41, in prepare_metadata_for_build_wheel
poetry = Factory().create_poetry(Path(".").resolve(), with_groups=False)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/private/var/folders/st/_0g2w6450zs5w9xts7h111f00000gn/T/pip-build-env-gq1yq4xp/overlay/lib/python3.11/site-packages/poetry/core/factory.py", line 58, in create_poetry
raise RuntimeError("The Poetry configuration is invalid:\n" + message)
RuntimeError: The Poetry configuration is invalid:
- [extras.pipfile_deprecated_finder.2] 'pip-shims<=0.3.4' does not match '^[a-zA-Z-_.0-9]+$'
[end of output]
note: This error originates from a subprocess, and is likely not a problem with pip.
error: metadata-generation-failed
× Encountered error while generating package metadata.
╰─> See above for output.
note: This is an issue with the package mentioned above, not pip.
hint: See above for details.
Traceback (most recent call last):
File "/Users/hayer/.virtualenvs/xlwings_311/lib/python3.11/site-packages/pre_commit/error_handler.py", line 73, in error_handler
yield
File "/Users/hayer/.virtualenvs/xlwings_311/lib/python3.11/site-packages/pre_commit/main.py", line 414, in main
return run(args.config, store, args)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/Users/hayer/.virtualenvs/xlwings_311/lib/python3.11/site-packages/pre_commit/commands/run.py", line 442, in run
install_hook_envs(to_install, store)
File "/Users/hayer/.virtualenvs/xlwings_311/lib/python3.11/site-packages/pre_commit/repository.py", line 248, in install_hook_envs
_hook_install(hook)
File "/Users/hayer/.virtualenvs/xlwings_311/lib/python3.11/site-packages/pre_commit/repository.py", line 95, in _hook_install
lang.install_environment(
File "/Users/hayer/.virtualenvs/xlwings_311/lib/python3.11/site-packages/pre_commit/languages/python.py", line 214, in install_environment
lang_base.setup_cmd(prefix, install_cmd)
File "/Users/hayer/.virtualenvs/xlwings_311/lib/python3.11/site-packages/pre_commit/lang_base.py", line 86, in setup_cmd
cmd_output_b(*cmd, cwd=prefix.prefix_dir, **kwargs)
File "/Users/hayer/.virtualenvs/xlwings_311/lib/python3.11/site-packages/pre_commit/util.py", line 110, in cmd_output_b
raise CalledProcessError(returncode, cmd, stdout_b, stderr_b)
pre_commit.util.CalledProcessError: command: ('/Users/hayer/.cache/pre-commit/repojetja5sx/py_env-python3/bin/python', '-mpip', 'install', '.')
return code: 1
stdout:
Processing /Users/hayer/.cache/pre-commit/repojetja5sx
Installing build dependencies: started
Installing build dependencies: finished with status 'done'
Getting requirements to build wheel: started
Getting requirements to build wheel: finished with status 'done'
Preparing metadata (pyproject.toml): started
Preparing metadata (pyproject.toml): finished with status 'error'
stderr:
error: subprocess-exited-with-error
× Preparing metadata (pyproject.toml) did not run successfully.
│ exit code: 1
╰─> [17 lines of output]
Traceback (most recent call last):
File "/Users/hayer/.cache/pre-commit/repojetja5sx/py_env-python3/lib/python3.11/site-packages/pip/_vendor/pyproject_hooks/_in_process/_in_process.py", line 353, in <module>
main()
File "/Users/hayer/.cache/pre-commit/repojetja5sx/py_env-python3/lib/python3.11/site-packages/pip/_vendor/pyproject_hooks/_in_process/_in_process.py", line 335, in main
json_out['return_val'] = hook(**hook_input['kwargs'])
^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/Users/hayer/.cache/pre-commit/repojetja5sx/py_env-python3/lib/python3.11/site-packages/pip/_vendor/pyproject_hooks/_in_process/_in_process.py", line 149, in prepare_metadata_for_build_wheel
return hook(metadata_directory, config_settings)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/private/var/folders/st/_0g2w6450zs5w9xts7h111f00000gn/T/pip-build-env-gq1yq4xp/overlay/lib/python3.11/site-packages/poetry/core/masonry/api.py", line 41, in prepare_metadata_for_build_wheel
poetry = Factory().create_poetry(Path(".").resolve(), with_groups=False)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/private/var/folders/st/_0g2w6450zs5w9xts7h111f00000gn/T/pip-build-env-gq1yq4xp/overlay/lib/python3.11/site-packages/poetry/core/factory.py", line 58, in create_poetry
raise RuntimeError("The Poetry configuration is invalid:\n" + message)
RuntimeError: The Poetry configuration is invalid:
- [extras.pipfile_deprecated_finder.2] 'pip-shims<=0.3.4' does not match '^[a-zA-Z-_.0-9]+$'
[end of output]
note: This error originates from a subprocess, and is likely not a problem with pip.
error: metadata-generation-failed
× Encountered error while generating package metadata.
╰─> See above for output.
note: This is an issue with the package mentioned above, not pip.
hint: See above for details.
```
|
closed
|
2023-05-29T13:19:54Z
|
2023-05-30T08:01:53Z
|
https://github.com/xlwings/xlwings/issues/2273
|
[] |
Jeroendevr
| 2
|
microsoft/nni
|
machine-learning
| 5,493
|
NNI gets stuck in WAITING state after a random number of successful trials
|
**Describe the issue**:
I am currently training an ML model (transformer) and I use NNI to tune the hyperparameters. The problem is that NNI gets stuck after a random number of successful trials. It seems to be unable to create the next upcoming trial (which would be the trial GPJNB). Rather, it throws an error because it can't find the trial it was unable to create in the first place (see figure with nnimanager.log).
**Environment**:
- NNI version: 2.9
- Training service (local|remote|pai|aml|etc): I think it's remote, see explanation below:
I am working with Windows Server 2019 in a virtual environment provided by my employer. But the model training is done on a computer cluster, not on my computer. I am using vpn connection to access this computer cluster. The operating system of the cluster is linux (CentOS Linux release 7.9.2009) and I use VSC as IDE (don't know if this is relevant). Submitting jobs works with slurm on this cluster.
- Python version: 3.8.3
- PyTorch/TensorFlow version: 2.9.1
- Is conda/virtualenv/venv used?: YES
- Is running in Docker?: No
**Log message**:
- nnimanager.log:
```
[2023-03-24 16:15:29] INFO (main) Start NNI manager
[2023-03-24 16:15:29] INFO (NNIDataStore) Datastore initialization done
[2023-03-24 16:15:29] INFO (RestServer) Starting REST server at port 8080, URL prefix: "/"
[2023-03-24 16:15:29] INFO (RestServer) REST server started.
[2023-03-24 16:15:29] INFO (NNIManager) Starting experiment: wy3mjz54
[2023-03-24 16:15:29] INFO (NNIManager) Setup training service...
[2023-03-24 16:15:29] INFO (LocalTrainingService) Construct local machine training service.
[2023-03-24 16:15:29] INFO (NNIManager) Setup tuner...
[2023-03-24 16:15:29] INFO (NNIManager) Change NNIManager status from: INITIALIZED to: RUNNING
[2023-03-24 16:15:30] INFO (NNIManager) Add event listeners
[2023-03-24 16:15:30] INFO (LocalTrainingService) Run local machine training service.
[2023-03-24 16:15:30] INFO (NNIManager) NNIManager received command from dispatcher: ID,
[2023-03-24 16:15:30] INFO (NNIManager) NNIManager received command from dispatcher: TR, {"parameter_id": 0, "parameter_source": "algorithm", "parameters": {"lr": 0.0008, "dr": 0.15, "bs": 16, "n_block": 3, "n_head": 4, "hidden_size": 128}, "parameter_index": 0}
[2023-03-24 16:15:35] INFO (NNIManager) submitTrialJob: form: {
sequenceId: 0,
hyperParameters: {
value: '{"parameter_id": 0, "parameter_source": "algorithm", "parameters": {"lr": 0.0008, "dr": 0.15, "bs": 16, "n_block": 3, "n_head": 4, "hidden_size": 128}, "parameter_index": 0}',
index: 0
},
placementConstraint: { type: 'None', gpus: [] }
}
[2023-03-24 16:15:45] INFO (NNIManager) Trial job qXJxV status changed from WAITING to RUNNING
[2023-03-24 16:15:55] INFO (main) Start NNI manager
[2023-03-24 16:15:55] INFO (NNIDataStore) Datastore initialization done
[2023-03-24 16:15:55] INFO (RestServer) Starting REST server at port 8111, URL prefix: "/"
[2023-03-24 16:15:55] INFO (RestServer) REST server started.
[2023-03-24 16:15:56] INFO (NNIManager) Resuming experiment: wy3mjz54
[2023-03-24 16:15:56] INFO (NNIManager) Setup training service...
[2023-03-24 16:15:56] INFO (LocalTrainingService) Construct local machine training service.
[2023-03-24 16:15:56] INFO (NNIManager) Change NNIManager status from: INITIALIZED to: VIEWED
[2023-03-25 00:29:26] INFO (NNIManager) Trial job qXJxV status changed from RUNNING to SUCCEEDED
[2023-03-25 00:29:26] INFO (NNIManager) NNIManager received command from dispatcher: TR, {"parameter_id": 1, "parameter_source": "algorithm", "parameters": {"lr": 0.0008, "dr": 0.15, "bs": 16, "n_block": 3, "n_head": 4, "hidden_size": 256}, "parameter_index": 0}
[2023-03-25 00:29:31] INFO (NNIManager) submitTrialJob: form: {
sequenceId: 1,
hyperParameters: {
value: '{"parameter_id": 1, "parameter_source": "algorithm", "parameters": {"lr": 0.0008, "dr": 0.15, "bs": 16, "n_block": 3, "n_head": 4, "hidden_size": 256}, "parameter_index": 0}',
index: 0
},
placementConstraint: { type: 'None', gpus: [] }
}
[2023-03-25 00:29:36] INFO (NNIManager) Trial job W4vPm status changed from WAITING to RUNNING
[2023-03-25 08:46:49] INFO (NNIManager) Trial job W4vPm status changed from RUNNING to FAILED
[2023-03-25 08:46:49] INFO (NNIManager) NNIManager received command from dispatcher: TR, {"parameter_id": 2, "parameter_source": "algorithm", "parameters": {"lr": 0.0008, "dr": 0.15, "bs": 16, "n_block": 3, "n_head": 4, "hidden_size": 512}, "parameter_index": 0}
[2023-03-25 08:46:54] INFO (NNIManager) submitTrialJob: form: {
sequenceId: 2,
hyperParameters: {
value: '{"parameter_id": 2, "parameter_source": "algorithm", "parameters": {"lr": 0.0008, "dr": 0.15, "bs": 16, "n_block": 3, "n_head": 4, "hidden_size": 512}, "parameter_index": 0}',
index: 0
},
placementConstraint: { type: 'None', gpus: [] }
}
[2023-03-25 08:46:59] INFO (NNIManager) Trial job eeLHB status changed from WAITING to RUNNING
[2023-03-25 17:13:02] INFO (NNIManager) Trial job eeLHB status changed from RUNNING to SUCCEEDED
[2023-03-25 17:13:02] INFO (NNIManager) NNIManager received command from dispatcher: TR, {"parameter_id": 3, "parameter_source": "algorithm", "parameters": {"lr": 0.0008, "dr": 0.15, "bs": 16, "n_block": 3, "n_head": 8, "hidden_size": 128}, "parameter_index": 0}
[2023-03-25 17:13:07] INFO (NNIManager) submitTrialJob: form: {
sequenceId: 3,
hyperParameters: {
value: '{"parameter_id": 3, "parameter_source": "algorithm", "parameters": {"lr": 0.0008, "dr": 0.15, "bs": 16, "n_block": 3, "n_head": 8, "hidden_size": 128}, "parameter_index": 0}',
index: 0
},
placementConstraint: { type: 'None', gpus: [] }
}
[2023-03-25 17:13:12] INFO (NNIManager) Trial job pFpV9 status changed from WAITING to RUNNING
[2023-03-26 09:55:19] INFO (NNIManager) Trial job pFpV9 status changed from RUNNING to SUCCEEDED
[2023-03-26 09:55:19] INFO (NNIManager) NNIManager received command from dispatcher: TR, {"parameter_id": 4, "parameter_source": "algorithm", "parameters": {"lr": 0.0008, "dr": 0.15, "bs": 16, "n_block": 3, "n_head": 8, "hidden_size": 256}, "parameter_index": 0}
[2023-03-26 09:55:24] INFO (NNIManager) submitTrialJob: form: {
sequenceId: 4,
hyperParameters: {
value: '{"parameter_id": 4, "parameter_source": "algorithm", "parameters": {"lr": 0.0008, "dr": 0.15, "bs": 16, "n_block": 3, "n_head": 8, "hidden_size": 256}, "parameter_index": 0}',
index: 0
},
placementConstraint: { type: 'None', gpus: [] }
}
[2023-03-26 09:55:29] INFO (NNIManager) Trial job YQvXf status changed from WAITING to RUNNING
[2023-03-27 02:07:54] INFO (NNIManager) Trial job YQvXf status changed from RUNNING to FAILED
[2023-03-27 02:07:54] INFO (NNIManager) NNIManager received command from dispatcher: TR, {"parameter_id": 5, "parameter_source": "algorithm", "parameters": {"lr": 0.0008, "dr": 0.15, "bs": 16, "n_block": 3, "n_head": 8, "hidden_size": 512}, "parameter_index": 0}
[2023-03-27 02:07:59] INFO (NNIManager) submitTrialJob: form: {
sequenceId: 5,
hyperParameters: {
value: '{"parameter_id": 5, "parameter_source": "algorithm", "parameters": {"lr": 0.0008, "dr": 0.15, "bs": 16, "n_block": 3, "n_head": 8, "hidden_size": 512}, "parameter_index": 0}',
index: 0
},
placementConstraint: { type: 'None', gpus: [] }
}
[2023-03-27 02:08:04] INFO (NNIManager) Trial job OiNAq status changed from WAITING to RUNNING
[2023-03-27 18:10:52] INFO (NNIManager) Trial job OiNAq status changed from RUNNING to FAILED
[2023-03-27 18:10:52] INFO (NNIManager) NNIManager received command from dispatcher: TR, {"parameter_id": 6, "parameter_source": "algorithm", "parameters": {"lr": 0.0008, "dr": 0.15, "bs": 16, "n_block": 3, "n_head": 16, "hidden_size": 128}, "parameter_index": 0}
[2023-03-27 18:10:57] INFO (NNIManager) submitTrialJob: form: {
sequenceId: 6,
hyperParameters: {
value: '{"parameter_id": 6, "parameter_source": "algorithm", "parameters": {"lr": 0.0008, "dr": 0.15, "bs": 16, "n_block": 3, "n_head": 16, "hidden_size": 128}, "parameter_index": 0}',
index: 0
},
placementConstraint: { type: 'None', gpus: [] }
}
[2023-03-27 18:11:03] INFO (NNIManager) Trial job DApPH status changed from WAITING to RUNNING
[2023-03-29 01:42:11] INFO (NNIManager) Trial job DApPH status changed from RUNNING to SUCCEEDED
[2023-03-29 01:42:11] INFO (NNIManager) NNIManager received command from dispatcher: TR, {"parameter_id": 7, "parameter_source": "algorithm", "parameters": {"lr": 0.0008, "dr": 0.15, "bs": 16, "n_block": 3, "n_head": 16, "hidden_size": 256}, "parameter_index": 0}
[2023-03-29 01:42:16] INFO (NNIManager) submitTrialJob: form: {
sequenceId: 7,
hyperParameters: {
value: '{"parameter_id": 7, "parameter_source": "algorithm", "parameters": {"lr": 0.0008, "dr": 0.15, "bs": 16, "n_block": 3, "n_head": 16, "hidden_size": 256}, "parameter_index": 0}',
index: 0
},
placementConstraint: { type: 'None', gpus: [] }
}
[2023-03-29 17:14:03] ERROR (NNIRestHandler) Error: File not found: /gpfs0/home/user/nni-experiments/wy3mjz54/trials/GPJNB/stderr
at LocalTrainingService.getTrialFile (/gpfs0/home/user/.conda/envs/FOMO/lib/python3.10/site-packages/nni_node/training_service/local/localTrainingService.js:146:19)
at NNIManager.getTrialFile (/gpfs0/home/user/.conda/envs/FOMO/lib/python3.10/site-packages/nni_node/core/nnimanager.js:333:37)
at /gpfs0/home/user/.conda/envs/FOMO/lib/python3.10/site-packages/nni_node/rest_server/restHandler.js:284:29
at Layer.handle [as handle_request] (/gpfs0/home/user/.conda/envs/FOMO/lib/python3.10/site-packages/nni_node/node_modules/express/lib/router/layer.js:95:5)
at next (/gpfs0/home/user/.conda/envs/FOMO/lib/python3.10/site-packages/nni_node/node_modules/express/lib/router/route.js:137:13)
at Route.dispatch (/gpfs0/home/user/.conda/envs/FOMO/lib/python3.10/site-packages/nni_node/node_modules/express/lib/router/route.js:112:3)
at Layer.handle [as handle_request] (/gpfs0/home/user/.conda/envs/FOMO/lib/python3.10/site-packages/nni_node/node_modules/express/lib/router/layer.js:95:5)
at /gpfs0/home/user/.conda/envs/FOMO/lib/python3.10/site-packages/nni_node/node_modules/express/lib/router/index.js:281:22
at param (/gpfs0/home/user/.conda/envs/FOMO/lib/python3.10/site-packages/nni_node/node_modules/express/lib/router/index.js:360:14)
at param (/gpfs0/home/user/.conda/envs/FOMO/lib/python3.10/site-packages/nni_node/node_modules/express/lib/router/index.js:371:14)
```
- dispatcher.log:
```
[2023-03-24 16:15:30] INFO (nni.tuner.gridsearch/MainThread) Ignored optimize_mode "maximize"
[2023-03-24 16:15:30] INFO (nni.runtime.msg_dispatcher_base/MainThread) Dispatcher started
[2023-03-24 16:15:30] INFO (nni.tuner.gridsearch/Thread-1 (command_queue_worker)) Grid initialized, size: (1×1×2×3×3×3) = 54
```
- nnictl stdout and stderr: For the respective trial (GPJNB), these files have not been created yet, as the status remains WAITING.
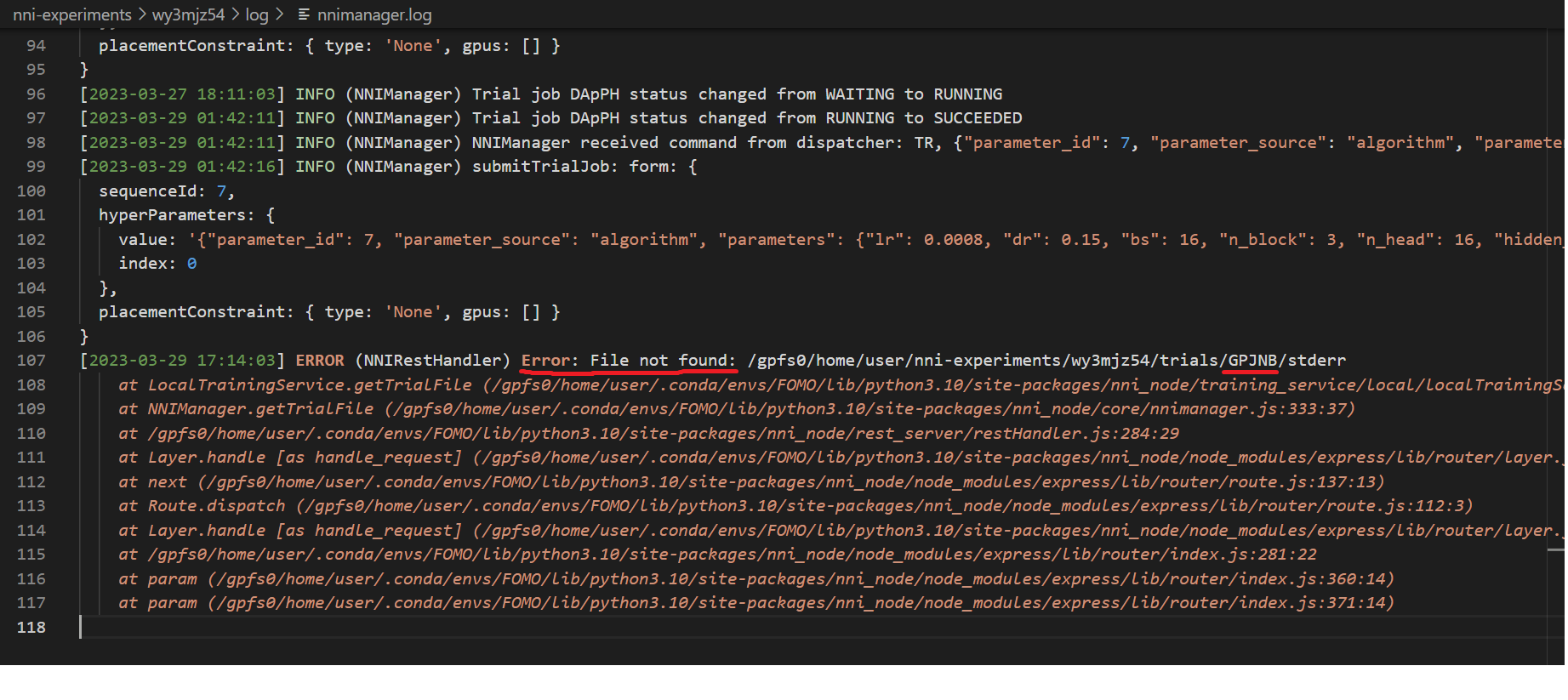
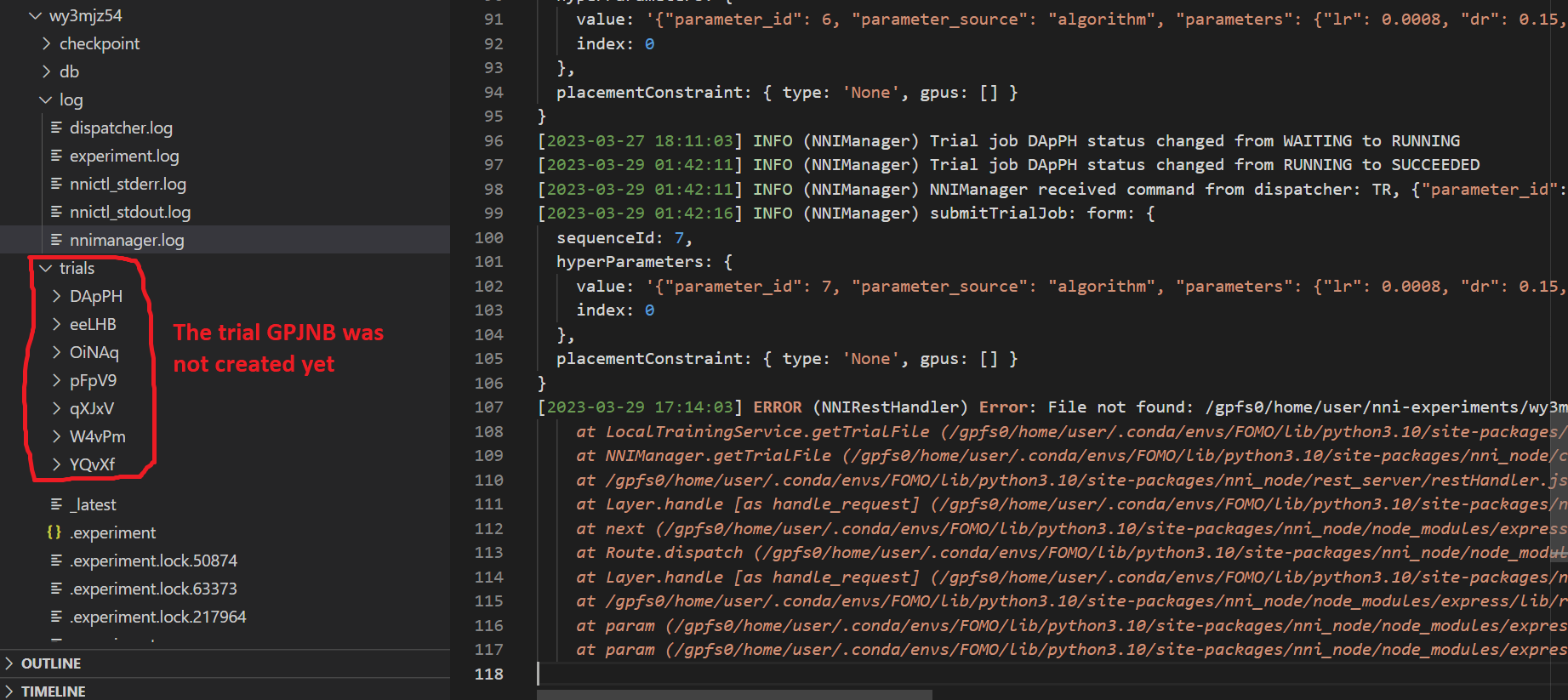
|
open
|
2023-03-29T16:36:58Z
|
2023-05-08T02:17:37Z
|
https://github.com/microsoft/nni/issues/5493
|
[] |
Oxid1
| 4
|
iMerica/dj-rest-auth
|
rest-api
| 523
|
Why commit=False in RegisterSerialzer?
|
Hi,
I use Django Allauth in my projects and wanted to extend my website with this rest endpoint implementation.
In /dj_rest_auth/registration/serialziers.py you set line 263 the commit option to False. Why?
```
def save(self, request):
adapter = get_adapter()
user = adapter.new_user(request)
self.cleaned_data = self.get_cleaned_data()
user = adapter.save_user(request, user, self, commit=False)
if "password1" in self.cleaned_data:
try:
adapter.clean_password(self.cleaned_data['password1'], user=user)
except DjangoValidationError as exc:
raise serializers.ValidationError(
detail=serializers.as_serializer_error(exc)
)
user.save()
self.custom_signup(request, user)
setup_user_email(request, user, [])
return user
```
This beaks my code, because in my custom adapter i do some actions with the user object, but if commit=False I can not add a foreign key to a new objects which is created in my custom adapter.
Thank you
|
open
|
2023-06-28T13:20:15Z
|
2023-09-08T22:47:47Z
|
https://github.com/iMerica/dj-rest-auth/issues/523
|
[] |
sowinski
| 1
|
suitenumerique/docs
|
django
| 434
|
Adapt export to new BlockNote.js version
|
## Feature Request
**Is your feature request related to a problem or unsupported use case? Please describe.**
With the version of BlockNote.js we can do exports from the client side.
**Describe the solution you'd like**
Adapt the export function so that it uses the BlockNote.js feature that works on the client side.
|
closed
|
2024-11-20T17:28:39Z
|
2025-01-28T12:36:05Z
|
https://github.com/suitenumerique/docs/issues/434
|
[
"frontend",
"editor"
] |
virgile-dev
| 0
|
Significant-Gravitas/AutoGPT
|
python
| 9,622
|
Implement popout text field on Agent Run page
|
The "New run" draft view has simple text inputs, which aren't great for long text inputs:
<img src="https://uploads.linear.app/a47946b5-12cd-4b3d-8822-df04c855879f/8b8c1083-e001-49f1-b90d-007976dc5120/e8e6c003-e3a2-42a8-aa32-26cd1454ff28?signature=eyJhbGciOiJIUzI1NiIsInR5cCI6IkpXVCJ9.eyJwYXRoIjoiL2E0Nzk0NmI1LTEyY2QtNGIzZC04ODIyLWRmMDRjODU1ODc5Zi84YjhjMTA4My1lMDAxLTQ5ZjEtYjkwZC0wMDc5NzZkYzUxMjAvZThlNmMwMDMtZTNhMi00MmE4LWFhMzItMjZjZDE0NTRmZjI4IiwiaWF0IjoxNzQxNzgyMzI4LCJleHAiOjMzMzEyMzQyMzI4fQ.RRu4OqWX-GdU7DgYDTPosnX4omtoSD01VSktpdls_ak " alt="image.png" width="1408" data-linear-height="565" />
To make this easier, add an "expand"/"pop out" button like node inputs have:
<img src="https://uploads.linear.app/a47946b5-12cd-4b3d-8822-df04c855879f/1a110b6b-ddfe-4354-b478-c5516b383bfe/d47c3a08-60b8-46c6-bcd4-67a100d623c3?signature=eyJhbGciOiJIUzI1NiIsInR5cCI6IkpXVCJ9.eyJwYXRoIjoiL2E0Nzk0NmI1LTEyY2QtNGIzZC04ODIyLWRmMDRjODU1ODc5Zi8xYTExMGI2Yi1kZGZlLTQzNTQtYjQ3OC1jNTUxNmIzODNiZmUvZDQ3YzNhMDgtNjBiOC00NmM2LWJjZDQtNjdhMTAwZDYyM2MzIiwiaWF0IjoxNzQxNzgyMzI4LCJleHAiOjMzMzEyMzQyMzI4fQ.ML30IjcJEN3B818OjExkGF-4O1Xly9h2zA_E1W5b_jI " alt="image.png" width="829" data-linear-height="960" />
This should be implemented as a feature on the `ui/Input` component, with an `expandable` property to enable it.
|
closed
|
2025-03-12T12:25:28Z
|
2025-03-18T15:33:28Z
|
https://github.com/Significant-Gravitas/AutoGPT/issues/9622
|
[
"UI",
"Feature",
"platform/frontend"
] |
Pwuts
| 0
|
awtkns/fastapi-crudrouter
|
fastapi
| 122
|
Support for Async SQLAlchemy
|
Hi everyone,
Is there any perspective of support for async SQLAlchemy?
Im using AssyncSession.
Thanks
|
open
|
2021-12-03T16:57:45Z
|
2023-05-10T08:43:38Z
|
https://github.com/awtkns/fastapi-crudrouter/issues/122
|
[
"enhancement"
] |
jrlopes2005
| 8
|
itamarst/eliot
|
numpy
| 122
|
Create explicit IDestination interface
|
It would be good to have explicit definition of destinations, and a generic test or two.
|
open
|
2014-10-21T18:29:46Z
|
2018-09-22T20:59:15Z
|
https://github.com/itamarst/eliot/issues/122
|
[
"documentation",
"API enhancement"
] |
itamarst
| 0
|
pennersr/django-allauth
|
django
| 3,265
|
ACCOUNT_EMAIL_CONFIRMATION_COOLDOWN creates failing tests
|
I upgraded all my dependencies today, and this setting caused me quite some headache.
I have some custom sign-up logic that I test quite extensively, and because of this setting, random tests kept failing, but not when executing them individually.
Maybe there is a clean way to disable this setting when testing, or a hint in the documentation? 🤔
|
closed
|
2023-02-17T15:09:14Z
|
2023-06-20T18:41:30Z
|
https://github.com/pennersr/django-allauth/issues/3265
|
[] |
Hafnernuss
| 1
|
junyanz/pytorch-CycleGAN-and-pix2pix
|
deep-learning
| 837
|
Generating images of same file type as input
|
Hello, I am using tiff images in the training of my pix2pix model. However, during the training and testing, the images generated are in the png format instead of tiff. As a result, the generated png image is completely black. Is there any way I can modify the code to generate images in a tiff format? Thank you!
|
closed
|
2019-11-12T11:22:51Z
|
2019-11-17T02:57:29Z
|
https://github.com/junyanz/pytorch-CycleGAN-and-pix2pix/issues/837
|
[] |
liuhh02
| 9
|
OpenInterpreter/open-interpreter
|
python
| 584
|
I started with interpreter --fast but it cost me a lot and it's very annoying
|
### Describe the bug
I started with interpreter --fast but it cost me a lot and it's really annoying ;(
please everytime we start a new session, write down everything
Model set to GPT-4
Open Interpreter will require approval before running code.
Use interpreter -y to bypass this.
**ADD SOMETHING LIKE THIS.**
To use GPT-3.5 Turbo: interpreter -- fast
To use locally: --local
Press CTRL-C to exit.
I'm very angry to pay that much for nothing. If i didn't go to my billing account to check i would finish the day with a $100 bill.
I'm very upset right now.
### Reproduce
interpreter --fast
### Expected behavior
- You should confirm what we use GPT-3.5 Turbo or Locally
### Screenshots
_No response_
### Open Interpreter version
latest
### Python version
latest
### Operating System name and version
Windows 10
### Additional context
Again. I'm very angry to pay that much for nothing. If i didn't go to my billing account to check i would finish the day with a $100 bill.
I'm very upset right now.
All these infos on how to use interpreter should be in the principal Readme page on this repo.
The first time days ago on using it, cost me a lot.
|
closed
|
2023-10-04T06:09:53Z
|
2023-10-30T00:04:08Z
|
https://github.com/OpenInterpreter/open-interpreter/issues/584
|
[
"Bug"
] |
onigetoc
| 13
|
Esri/arcgis-python-api
|
jupyter
| 1,408
|
Current ArcPy version is unbuildable from Anaconda distribution
|
**Describe the bug**
Utilizing the standard command `conda install -c esri arcpy` in a new, empty, conda environment produces a build error which aborts the installation.
**To Reproduce**
Steps to reproduce the behavior:
```
conda create -n clean_env
y
conda activate clean_env
conda install -c esri arcpy
y
```
error:
```
ERROR conda.core.link:_execute(740): An error occurred while installing package 'esri::jupyter_contrib_nbextensions-0.5.1-py_24'.
Rolling back transaction: done
LinkError: post-link script failed for package esri::jupyter_contrib_nbextensions-0.5.1-py_24
location of failed script: E:\Anaconda3\envs\clean_env\Scripts\.jupyter_contrib_nbextensions-post-link.bat
==> script messages <==
<None>
==> script output <==
stdout:
stderr: Traceback (most recent call last):
File "E:\Anaconda3\envs\clean_env\Scripts\jupyter-contrib-nbextension-script.py", line 5, in <module>
from jupyter_contrib_nbextensions.application import main
File "E:\Anaconda3\envs\clean_env\lib\site-packages\jupyter_contrib_nbextensions\application.py", line 15, in <module>
from jupyter_contrib_nbextensions.install import (
File "E:\Anaconda3\envs\clean_env\lib\site-packages\jupyter_contrib_nbextensions\install.py", line 12, in <module>
import latex_envs
File "E:\Anaconda3\envs\clean_env\lib\site-packages\latex_envs\__init__.py", line 3, in <module>
from . import latex_envs
File "E:\Anaconda3\envs\clean_env\lib\site-packages\latex_envs\latex_envs.py", line 20, in <module>
from nbconvert.exporters.exporter import Exporter
File "E:\Anaconda3\envs\clean_env\lib\site-packages\nbconvert\__init__.py", line 4, in <module>
from .exporters import *
File "E:\Anaconda3\envs\clean_env\lib\site-packages\nbconvert\exporters\__init__.py", line 3, in <module>
from .html import HTMLExporter
File "E:\Anaconda3\envs\clean_env\lib\site-packages\nbconvert\exporters\html.py", line 12, in <module>
from jinja2 import contextfilter
ImportError: cannot import name 'contextfilter' from 'jinja2' (E:\Anaconda3\envs\clean_env\lib\site-packages\jinja2\__init__.py)
return code: 1
()
```
**Screenshots**


**Expected behavior**
The build to complete successfully.
**Platform (please complete the following information):**
- OS: Windows 11
- Anaconda 2022.10
- arcgis-2.0.1-py39_2826
- arcgispro-3.0-0
- arcpy-3.0-py39_arcgispro_36056
**Additional context**
|
closed
|
2022-12-30T16:00:07Z
|
2023-08-08T14:50:42Z
|
https://github.com/Esri/arcgis-python-api/issues/1408
|
[
"bug"
] |
FeralCatColonist
| 6
|
Skyvern-AI/skyvern
|
api
| 1,540
|
Why does "Wokflow" in the localhost dashboard have 'Max Retries' while 'Task' does not?
|
Why does "Wokflow" in the localhost dashboard have 'Max Retries' while 'Task' does not? #1540
|
open
|
2025-01-12T13:55:09Z
|
2025-01-13T00:12:01Z
|
https://github.com/Skyvern-AI/skyvern/issues/1540
|
[] |
computer2s
| 1
|
strawberry-graphql/strawberry
|
graphql
| 3,678
|
handle_ping when client is disconnected raises an uncatched WebSocketDisconnect (sometimes)
|
Steps to repdroduce:
- Add a async.sleep(10s) to [handle_ping](https://github.com/strawberry-graphql/strawberry/blob/main/strawberry/subscriptions/protocols/graphql_transport_ws/handlers.py#L206)
- Open websocket
- wait for a ping (async.sleep(10s) is now running/blocking)
- disconnect client
- server tries to send a pong but websocket is disconnected and throws a WebSocketDisconnect (but not always???)
This looks like a racecondition, and I can't make a failing reliable example -.-'
|
closed
|
2024-10-23T07:00:32Z
|
2025-03-20T15:56:54Z
|
https://github.com/strawberry-graphql/strawberry/issues/3678
|
[
"bug"
] |
Speedy1991
| 3
|
autogluon/autogluon
|
data-science
| 4,466
|
No matching distribution found for numpy<3.0.0,>=2.0.0
|
System configuration: python3.8
Installing build dependencies ... error
error: subprocess-exited-with-error
× pip subprocess to install build dependencies did not run successfully.
│ exit code: 1
╰─> [74 lines of output]
Looking in indexes: https://repo.huaweicloud.com/repository/pypi/simple/
Ignoring numpy: markers 'python_version >= "3.9"' don't match your environment
Collecting setuptools
Downloading https://repo.huaweicloud.com/repository/pypi/packages/cb/9c/9ad11ac06b97e55ada655f8a6bea9d1d3f06e120b178cd578d80e558191d/setuptools-74.1.2-py3-none-any.whl (1.3 MB)
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 1.3/1.3 MB 38.3 MB/s eta 0:00:00
Collecting cython<3.0,>=0.25
Downloading https://repo.huaweicloud.com/repository/pypi/packages/10/46/9a1b428442d09a6496d56e913ff4c3f4a1db119adc45d45ad2655bcb2a68/Cython-0.29.37-cp38-cp38-manylinux_2_17_x86_64.manylinux2014_x86_64.manylinux_2_24_x86_64.whl (1.9 MB)
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 1.9/1.9 MB 37.2 MB/s eta 0:00:00
Collecting cymem<2.1.0,>=2.0.2
Downloading https://repo.huaweicloud.com/repository/pypi/packages/5f/70/b9945a7918d467c6c7112f6e20176d4f41b89d7ba0b590015b4cb62fb23d/cymem-2.0.8-cp38-cp38-manylinux_2_17_x86_64.manylinux2014_x86_64.whl (46 kB)
Collecting preshed<3.1.0,>=3.0.2
Downloading https://repo.huaweicloud.com/repository/pypi/packages/aa/7c/f36a498cb114765d0ecec946e6be46d2aadaea19317a1a1828e4aa2383d8/preshed-3.0.9-cp38-cp38-manylinux_2_5_x86_64.manylinux1_x86_64.manylinux_2_17_x86_64.manylinux2014_x86_64.whl (154 kB)
Collecting murmurhash<1.1.0,>=0.28.0
Downloading https://repo.huaweicloud.com/repository/pypi/packages/f9/63/49e1eda3c610f49e5d3062e44ed27751f33ca8c4087b100b78b141201a6a/murmurhash-1.0.10-cp38-cp38-manylinux_2_5_x86_64.manylinux1_x86_64.manylinux_2_17_x86_64.manylinux2014_x86_64.whl (29 kB)
Collecting thinc<8.4.0,>=8.3.0
Downloading https://repo.huaweicloud.com/repository/pypi/packages/15/0a/250f7fa34632616bb4fc37decbc46ed09243bade708968dc869d8a4c144b/thinc-8.3.0.tar.gz (193 kB)
Installing build dependencies: started
Installing build dependencies: finished with status 'error'
error: subprocess-exited-with-error
× pip subprocess to install build dependencies did not run successfully.
│ exit code: 1
╰─> [41 lines of output]
Looking in indexes: https://repo.huaweicloud.com/repository/pypi/simple/
Ignoring numpy: markers 'python_version >= "3.9"' don't match your environment
Collecting setuptools
Using cached https://repo.huaweicloud.com/repository/pypi/packages/cb/9c/9ad11ac06b97e55ada655f8a6bea9d1d3f06e120b178cd578d80e558191d/setuptools-74.1.2-py3-none-any.whl (1.3 MB)
Collecting cython<3.0,>=0.25
Using cached https://repo.huaweicloud.com/repository/pypi/packages/10/46/9a1b428442d09a6496d56e913ff4c3f4a1db119adc45d45ad2655bcb2a68/Cython-0.29.37-cp38-cp38-manylinux_2_17_x86_64.manylinux2014_x86_64.manylinux_2_24_x86_64.whl (1.9 MB)
Collecting murmurhash<1.1.0,>=1.0.2
Using cached https://repo.huaweicloud.com/repository/pypi/packages/f9/63/49e1eda3c610f49e5d3062e44ed27751f33ca8c4087b100b78b141201a6a/murmurhash-1.0.10-cp38-cp38-manylinux_2_5_x86_64.manylinux1_x86_64.manylinux_2_17_x86_64.manylinux2014_x86_64.whl (29 kB)
Collecting cymem<2.1.0,>=2.0.2
Using cached https://repo.huaweicloud.com/repository/pypi/packages/5f/70/b9945a7918d467c6c7112f6e20176d4f41b89d7ba0b590015b4cb62fb23d/cymem-2.0.8-cp38-cp38-manylinux_2_17_x86_64.manylinux2014_x86_64.whl (46 kB)
Collecting preshed<3.1.0,>=3.0.2
Using cached https://repo.huaweicloud.com/repository/pypi/packages/aa/7c/f36a498cb114765d0ecec946e6be46d2aadaea19317a1a1828e4aa2383d8/preshed-3.0.9-cp38-cp38-manylinux_2_5_x86_64.manylinux1_x86_64.manylinux_2_17_x86_64.manylinux2014_x86_64.whl (154 kB)
Collecting blis<1.1.0,>=1.0.0
Downloading https://repo.huaweicloud.com/repository/pypi/packages/b6/0b/a73be025d991e8795626a830314984f7bda5de440e80b72a53bc316bb870/blis-1.0.0.tar.gz (3.6 MB)
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 3.6/3.6 MB 59.9 MB/s eta 0:00:00
Installing build dependencies: started
Installing build dependencies: finished with status 'error'
error: subprocess-exited-with-error
× pip subprocess to install build dependencies did not run successfully.
│ exit code: 1
╰─> [9 lines of output]
Looking in indexes: https://repo.huaweicloud.com/repository/pypi/simple/
Collecting setuptools
Using cached https://repo.huaweicloud.com/repository/pypi/packages/cb/9c/9ad11ac06b97e55ada655f8a6bea9d1d3f06e120b178cd578d80e558191d/setuptools-74.1.2-py3-none-any.whl (1.3 MB)
Collecting cython>=0.25
Downloading https://repo.huaweicloud.com/repository/pypi/packages/b2/52/eda119f98071ccde04a9a1c9c9a18fd6def025651c9d0cd01ad51d0dba36/Cython-3.0.11-cp38-cp38-manylinux_2_17_x86_64.manylinux2014_x86_64.whl (3.6 MB)
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 3.6/3.6 MB 22.3 MB/s eta 0:00:00
ERROR: Ignored the following versions that require a different python version: 1.25.0 Requires-Python >=3.9; 1.25.1 Requires-Python >=3.9; 1.25.2 Requires-Python >=3.9; 1.26.0 Requires-Python <3.13,>=3.9; 1.26.1 Requires-Python <3.13,>=3.9; 1.26.2 Requires-Python >=3.9; 1.26.3 Requires-Python >=3.9; 1.26.4 Requires-Python >=3.9; 2.0.0 Requires-Python >=3.9; 2.0.1 Requires-Python >=3.9; 2.0.2 Requires-Python >=3.9; 2.1.0 Requires-Python >=3.10; 2.1.0rc1 Requires-Python >=3.10; 2.1.1 Requires-Python >=3.10
ERROR: Could not find a version that satisfies the requirement numpy<3.0.0,>=2.0.0 (from versions: 1.3.0, 1.4.1, 1.5.0, 1.5.1, 1.6.0, 1.6.1, 1.6.2, 1.7.0, 1.7.1, 1.7.2, 1.8.0, 1.8.1, 1.8.2, 1.9.0, 1.9.1, 1.9.2, 1.9.3, 1.10.0.post2, 1.10.1, 1.10.2, 1.10.4, 1.11.0, 1.11.1, 1.11.2, 1.11.3, 1.12.0, 1.12.1, 1.13.0, 1.13.1, 1.13.3, 1.14.0, 1.14.1, 1.14.2, 1.14.3, 1.14.4, 1.14.5, 1.14.6, 1.15.0, 1.15.1, 1.15.2, 1.15.3, 1.15.4, 1.16.0, 1.16.1, 1.16.2, 1.16.3, 1.16.4, 1.16.5, 1.16.6, 1.17.0, 1.17.1, 1.17.2, 1.17.3, 1.17.4, 1.17.5, 1.18.0, 1.18.1, 1.18.2, 1.18.3, 1.18.4, 1.18.5, 1.19.0, 1.19.1, 1.19.2, 1.19.3, 1.19.4, 1.19.5, 1.20.0, 1.20.1, 1.20.2, 1.20.3, 1.21.0, 1.21.1, 1.21.2, 1.21.3, 1.21.4, 1.21.5, 1.21.6, 1.22.0, 1.22.1, 1.22.2, 1.22.3, 1.22.4, 1.23.0, 1.23.1, 1.23.2, 1.23.3, 1.23.4, 1.23.5, 1.24.0, 1.24.1, 1.24.2, 1.24.3, 1.24.4)
ERROR: No matching distribution found for numpy<3.0.0,>=2.0.0
[end of output]
note: This error originates from a subprocess, and is likely not a problem with pip.
error: subprocess-exited-with-error
× pip subprocess to install build dependencies did not run successfully.
│ exit code: 1
╰─> See above for output.
note: This error originates from a subprocess, and is likely not a problem with pip.
[end of output]
note: This error originates from a subprocess, and is likely not a problem with pip.
error: subprocess-exited-with-error
× pip subprocess to install build dependencies did not run successfully.
│ exit code: 1
╰─> See above for output.
note: This error originates from a subprocess, and is likely not a problem with pip.
[end of output]
note: This error originates from a subprocess, and is likely not a problem with pip.
error: subprocess-exited-with-error
|
open
|
2024-09-12T08:46:53Z
|
2024-09-12T08:46:53Z
|
https://github.com/autogluon/autogluon/issues/4466
|
[
"bug: unconfirmed",
"Needs Triage"
] |
soaprockets
| 0
|
onnx/onnx
|
scikit-learn
| 6,670
|
linux aarch64 build is failing
|
# Bug Report
### Is the issue related to model conversion?
### Describe the bug
Currently we have build problems with linux aarch64 (memory exceptions / segmentations faults are appearing)
One of the latest successful runs was https://github.com/onnx/onnx/actions/runs/12775045059/job/35610508975#step:5:121
|
closed
|
2025-01-31T06:00:14Z
|
2025-02-14T04:01:30Z
|
https://github.com/onnx/onnx/issues/6670
|
[
"bug",
"contributions welcome"
] |
andife
| 2
|
rthalley/dnspython
|
asyncio
| 844
|
dns.resolver.Resolver fails with "dns.name.EmptyLabel: A DNS label is empty." when search list contains a suffix starting with a dot
|
**Describe the bug**
dns.resolver.Resolver fails with "dns.name.EmptyLabel: A DNS label is empty." when search list contains a suffix starting with a dot, for example .home as set by some SoHo routers.
**To Reproduce**
Python 3.8.10 (tags/v3.8.10:3d8993a, May 3 2021, 11:48:03) [MSC v.1928 64 bit (AMD64)] on win32
Type "help", "copyright", "credits" or "license" for more information.
>>> import dns.resolver
With WMI module installed:
>>> my_resolver = dns.resolver.Resolver()
Exception in thread Thread-1:
Traceback (most recent call last):
File "...\threading.py", line 932, in _bootstrap_inner
self.run()
File "...\win32util.py", line 50, in run
self.info.search = [dns.name.from_text(x) for x in
File "...\win32util.py", line 50, in <listcomp>
self.info.search = [dns.name.from_text(x) for x in
File "...\name.py", line 942, in from_text
raise EmptyLabel
dns.name.EmptyLabel: A DNS label is empty.
Without WMI module installed:
>>> my_resolver = dns.resolver.Resolver()
Traceback (most recent call last):
File "<stdin>", line 1, in <module>
File "...\resolver.py", line 756, in __init__
self.read_registry()
File "...\resolver.py", line 855, in read_registry
info = dns.win32util.get_dns_info()
File "...\win32util.py", line 235, in get_dns_info
return getter.get()
File "...\win32util.py", line 199, in get
self._config_fromkey(tcp_params, True)
File "...\win32util.py", line 138, in _config_fromkey
self._config_search(search)
File "...\win32util.py", line 97, in _config_search
s = dns.name.from_text(s)
File "...\name.py", line 942, in from_text
raise EmptyLabel
dns.name.EmptyLabel: A DNS label is empty.
DNS search list: ['example.com', '.home']
**Context (please complete the following information):**
- dnspython version 2.2.1
- Python version 3.8.10
- OS: Microsoft Windows [Version 10.0.19044.2006]
|
closed
|
2022-10-06T10:11:08Z
|
2022-10-06T12:52:45Z
|
https://github.com/rthalley/dnspython/issues/844
|
[
"Enhancement Request"
] |
krusch
| 3
|
plotly/dash-core-components
|
dash
| 222
|
Ability to set a maximum number of intervals?
|
As suggested by a community member:
> When I put it this way, the cleanest solution to my problem would be if `dcc.Interval()` supported the idea of counting a certain number of ticks and then stopping by itself, i.e. had something like a `max_intervals` parameter. I’m not sure if you have any other use-cases for that but if so, maybe it’s a feature suggestion?
Full discussion: https://community.plot.ly/t/should-i-use-an-interval-for-a-one-off-delay/11375/6
Seems like a pretty good idea, coul dbe useful for general delays and fits nicely into the existing component API
|
closed
|
2018-07-02T03:04:35Z
|
2018-07-12T10:44:04Z
|
https://github.com/plotly/dash-core-components/issues/222
|
[
"dash-type-enhancement",
"in progress",
"dash-meta-good_first_issue"
] |
chriddyp
| 1
|
ageitgey/face_recognition
|
machine-learning
| 1,267
|
train_object_detector
|
* face_recognition version: 1.4.0
* Python version: 2.7.16
* Operating System: Pi 4: Raspbian GNU/Linux 10 (buster)
### Description
Hello,
I would like to try using this for recognising individual animal faces. I found train_object_detector in dlib/examples and have been walking through that to re-train the object detector. I compiled the imglab tool and labeled my training images with examples of the faces I want to detect.
I am stuck on the next step. How can I compile train_object_detector? Could someone further explain these instructions to a noob?
“Returning to the present example program, we can compile it using cmake just as we did with the imglab tool. Once compiled, we can issue the command
./train_object_detector -tv mydataset.xml”
I tried to do this by both pointing to the examples folder containing the train_object_detector file and to the .cpp file itself as we did in the following instructions for the imglab tool:
“To create this annotated data you will need to use the imglab tool
included with dlib. It is located in the tools/imglab folder and can be compiled
using the following commands.
cd tools/imglab
mkdir build
cd build
cmake ..
cmake --build . --config Release”
### What I Did
cd /home/pi/dlib/dlib/examples/train_object_detector.cpp
mkdir build
cd build
cmake ..
cmake --build . --config Release
Result:
cd /home/pi/dlib/dlib/examples/train_object_detector.cpp
bash: cd: /home/pi/dlib/dlib/examples/train_object_detector.cpp: Not a directory
pi@raspberrypi:~/dlib/dlib/tools/train_object_detector/build $ mkdir build
pi@raspberrypi:~/dlib/dlib/tools/train_object_detector/build $ cd build
pi@raspberrypi:~/dlib/dlib/tools/train_object_detector/build/build $ cmake ..
CMake Error at CMakeLists.txt:43 (add_subdirectory):
add_subdirectory given source "../dlib" which is not an existing directory.
OpenCV not found, so we won't build the webcam_face_pose_ex example.
-- Configuring incomplete, errors occurred!
See also "/home/pi/dlib/dlib/tools/train_object_detector/build/CMakeFiles/CMakeOutput.log".
pi@raspberrypi:~/dlib/dlib/tools/train_object_detector/build/build $ cmake --build . --config Release
Error: could not load cache
Anyone know how I should compile it or in what location? I would think that you would want the object detector to be in the same directory of the annotated dataset mydataset.xml??
Thank you so much!
|
open
|
2021-01-15T16:53:15Z
|
2021-01-15T16:53:15Z
|
https://github.com/ageitgey/face_recognition/issues/1267
|
[] |
SquirrelMonkeyPrincess
| 0
|
widgetti/solara
|
flask
| 204
|
QST: solara usage with Treeview
|
First of all, thanks a lot for building this library. I really think the python community needs this.
I worked a lot with ipyvuetify, for my next application I am trying to used solara.
In this application I need to interact with a Treeview and I did not make it work with solara yet.
Basically I am trying to make something like this with `rw.Treeview` but I am missing the `on_event` function
```python
import ipyvuetify as v
items = [{'id' : 'a', 'name': 'A'}, {'id' : 'b', 'name': 'B'}]
class my_app(v.Col):
def __init__(self):
self.ipyvuetify_tree = v.Treeview(active = [], items = items, activatable=True)
self.text = v.Text(children = ['{}'])
self.ipyvuetify_tree.on_event('update:active', self.update_active)
super().__init__(
children=[self.ipyvuetify_tree, self.text]
)
def update_active(self, widget, event, data):
widget.active = data if data else []
self.text.children = [f'{data}']
```
Can you give me hints on how to use the `active` trait of `Treeview` ? Thanks !
|
closed
|
2023-07-10T15:53:17Z
|
2023-07-20T16:16:24Z
|
https://github.com/widgetti/solara/issues/204
|
[
"documentation"
] |
gab23r
| 4
|
browser-use/browser-use
|
python
| 330
|
browser-use + lightrag errors.
|
When I try to use browser-use and lightrag together, the only result I get is that it keeps crashing and I've been trying this for maybe 24 hours and I'm bored now. While browser-use works fine, when lightrag is included in the process it either gives an openai key error or another error. There are hundreds of errors and new ones are added as I fix them. I can't figure out whether my computer is too incompatible or I'm too far from artificial intelligence.
If you have a ready-made package, I would be very happy if you could leave a link to the package that works together properly.
|
open
|
2025-01-20T09:18:05Z
|
2025-01-29T18:02:11Z
|
https://github.com/browser-use/browser-use/issues/330
|
[] |
Kerimtunc
| 1
|
cobrateam/splinter
|
automation
| 1,036
|
(selenium) UserWarning: find_elements_by_css_selector is deprecated
|
Hello, didn't see an open issue on this so letting devs know about
`/usr/local/lib/python3.9/site-packages/selenium/webdriver/remote/webelement.py:502: UserWarning: find_elements_by_css_selector is deprecated. Please use find_elements(by=By.CSS_SELECTOR, value=css_selector) instead
warnings.warn("find_elements_by_css_selector is deprecated. Please use find_elements(by=By.CSS_SELECTOR, value=css_selector) instead")`
splinter==0.17.0
selenium==4.1.3
|
closed
|
2022-05-05T18:14:55Z
|
2022-05-05T18:43:09Z
|
https://github.com/cobrateam/splinter/issues/1036
|
[] |
walksonair
| 1
|
explosion/spaCy
|
machine-learning
| 13,112
|
Error Message while trying to use spaCy experimental package for CorefResolver
|
While using spaCy experimental package for CorefResolver, I am getting the following error, Any clue what package could be missing?
RegistryError: [E893] Could not find function 'spacy-experimental.Coref.v1' in function registry 'architectures'. If you're using a custom function, make sure the code is available. If the function is provided by a third-party package, e.g. spacy-transformers, make sure the package is installed in your environment.
Available names: spacy-legacy.CharacterEmbed.v1, spacy-legacy.EntityLinker.v1, spacy-legacy.HashEmbedCNN.v1, spacy-legacy.MaxoutWindowEncoder.v1, spacy-legacy.MishWindowEncoder.v1, spacy-legacy.MultiHashEmbed.v1, spacy-legacy.Tagger.v1, spacy-legacy.TextCatBOW.v1, spacy-legacy.TextCatCNN.v1, spacy-legacy.TextCatEnsemble.v1, spacy-legacy.Tok2Vec.v1, spacy-legacy.TransitionBasedParser.v1, spacy.CharacterEmbed.v2, spacy.EntityLinker.v2, spacy.HashEmbedCNN.v2, spacy.MaxoutWindowEncoder.v2, spacy.MishWindowEncoder.v2, spacy.MultiHashEmbed.v2, spacy.PretrainCharacters.v1, spacy.PretrainVectors.v1, spacy.SpanCategorizer.v1, spacy.Tagger.v2, spacy.TextCatBOW.v2, spacy.TextCatCNN.v2, spacy.TextCatEnsemble.v2, spacy.TextCatLowData.v1, spacy.Tok2Vec.v2, spacy.Tok2VecListener.v1, spacy.TorchBiLSTMEncoder.v1, spacy.TransitionBasedParser.v2
## My Environment
* Operating System: Windows 11 (22H2)
* Python Version Used: v3.11.15
* spaCy Version Used: v3.5.3
* Code Sinippet:
>from spacy_experimental.coref.coref_component import DEFAULT_COREF_MODEL
>from spacy_experimental.coref.coref_util import DEFAULT_CLUSTER_PREFIX
> doc = nlp("This is a sentence.")
> coref = nlp.add_pipe("experimental_coref")
|
closed
|
2023-11-07T04:14:39Z
|
2023-11-07T08:39:46Z
|
https://github.com/explosion/spaCy/issues/13112
|
[] |
kausikb
| 1
|
TheKevJames/coveralls-python
|
pytest
| 90
|
You have to provide either repo_token in .coveralls.yml, or launch via Travis
|
I get that here: https://travis-ci.org/ionelmc/python-hunter/jobs/81493516#L286
Anything obvious that I'm doing wrong there?
|
closed
|
2015-09-28T15:26:27Z
|
2015-10-03T22:20:35Z
|
https://github.com/TheKevJames/coveralls-python/issues/90
|
[] |
ionelmc
| 1
|
redis/redis-om-python
|
pydantic
| 15
|
Document querying
|
Add a Markdown document to docs/ with more in-depth querying examples.
|
open
|
2021-11-13T01:36:50Z
|
2021-11-13T01:36:50Z
|
https://github.com/redis/redis-om-python/issues/15
|
[] |
abrookins
| 0
|
PrefectHQ/prefect
|
automation
| 17,005
|
allow manual control of `task`s
|
### Describe the current behavior
Currently the only (official/documented) way to define a task is adding `@task` decorator to functions. For jobs that involve large numbers of long interwoven concurrent tasks, they are often wrongly nested together in the Prefect Cloud UI.

### Describe the proposed behavior
I'd like to have the ability to:
- manually define a task's parent, so I can choose where to nest a task
- even better: manually create an empty task, which acts as a logical group of smaller tasks. This wrapping task will end when no more subtasks are manually added to it.
### Example Use
```python
def parse_sitemap(self, response, **cb_kwargs):
group = new_task()
...
for listing in listings:
yield Request(... cb_kwargs={'group_id' = group.id})
@staticmethod
def get_listing_parent():
parameters = task_run.parameters
group_id= parameters["cb_kwargs"].get('group_id')
return group_id
@task(name="Parse listing", parent=get_listing_parent)
def parse_listing(self, response, **cb_kwargs):
...
```
### Additional context
_No response_
|
open
|
2025-02-06T08:58:35Z
|
2025-02-06T15:12:56Z
|
https://github.com/PrefectHQ/prefect/issues/17005
|
[
"enhancement"
] |
kent-ps
| 0
|
flairNLP/flair
|
nlp
| 2,966
|
Fine-tune on specific domain
|
Hi,
We chose to use Flair for our domain-specific NER model.
So far we got excellent results!
There is something that I would like to clarify, We are tagging data all the time, which means that every couple of weeks we get another batch of labeled data.
For the first batch, we used TransformerWordEmbeddings with "allenai/longformer-base-4096" because we have long sentences data and then fine_tune the "SequenceTagger"
After the first training, we wanted to train our model on the new batch of data
What would be better in this case?
load our current SequenceTagger model with `SequenceTagger.load()` and use fine_tune again?
or load our current SequenceTagger model with `SequenceTagger.load()` and use train function?
or maybe train all the data together on a new SequenceTagger with TransformerWordEmbeddings?
One more problem I had, when I tried to load my model and use fine_tune again I ran into a CUDA OUT OF MEMORY error
while when I used all the data together and train a new SequenceTagger the train finished successfully
any ideas why?
We use AWS sageMaker instance type: ml.p3.2xlarge
Thanks ahead for any help
This is my code for the loading and training
```
# 4. initialize fine-tuneable transformer embeddings
embeddings = TransformerWordEmbeddings(model="allenai/longformer-base-4096",
layers="-1",
subtoken_pooling="first",
fine_tune=True,
use_context=True,
allow_long_sentences=True
)
embeddings.max_subtokens_sequence_length = 4096
# 5. initialize bare-bones sequence tagger (no CRF, no RNN, no reprojection)
new_model = False
if new_model:
tagger = SequenceTagger(hidden_size=256,
embeddings=embeddings,
tag_dictionary=label_dict,
tag_type='ner',
use_crf=False,
use_rnn=False,
reproject_embeddings=False,
)
else:
tagger = SequenceTagger.load('round_7_full_address_fine_tune/final-model.pt')
tagger.embeddings.max_subtokens_sequence_length = 4096
# 6. initialize trainer
trainer = ModelTrainer(tagger, corpus)
model_path = f'round_{ROUND_NUMBER}_full_address{FINE_TUNED}_similars'
# 7. run fine-tuning
trainer.fine_tune(model_path,
learning_rate=1.0e-5,
mini_batch_size=2,
write_weights = True,
mini_batch_chunk_size=4, # remove this parameter to speed up computation if you have a big GPU
embeddings_storage_mode='none'
)
```
|
closed
|
2022-10-21T11:58:51Z
|
2023-01-17T18:42:03Z
|
https://github.com/flairNLP/flair/issues/2966
|
[
"question"
] |
shahafp
| 4
|
JaidedAI/EasyOCR
|
pytorch
| 731
|
finetuning easyocr using persian handwritten data
|
can easyocr be finetuned using persian handwritten data?
|
open
|
2022-05-19T08:41:07Z
|
2022-05-19T08:41:07Z
|
https://github.com/JaidedAI/EasyOCR/issues/731
|
[] |
Nadiam75
| 0
|
recommenders-team/recommenders
|
machine-learning
| 2,126
|
[BUG] Cornac BiVAE test failing due to csc_matrix attribute error
|
### Description
<!--- Describe your issue/bug/request in detail -->
```
E bivae = cornac.models.BiVAECF(
E k=LATENT_DIM,
E encoder_structure=ENCODER_DIMS,
E act_fn=ACT_FUNC,
E likelihood=LIKELIHOOD,
E n_epochs=NUM_EPOCHS,
E batch_size=BATCH_SIZE,
E learning_rate=LEARNING_RATE,
E seed=SEED,
E use_gpu=torch.cuda.is_available(),
E verbose=True
E )
E
E with Timer() as t:
E bivae.fit(train_set)
E print("Took *** seconds for training.".format(t))
E ------------------
E
E ----- stderr -----
E
0%| | 0/500 [00:00<?, ?it/s]
E ----- stderr -----
E
0%| | 0/500 [00:00<?, ?it/s]
E ----- stderr -----
E
E ------------------
E
E ---------------------------------------------------------------------------
E AttributeError Traceback (most recent call last)
E Cell In[6], line 15
E 1 bivae = cornac.models.BiVAECF(
E 2 k=LATENT_DIM,
E 3 encoder_structure=ENCODER_DIMS,
E (...)
E 11 verbose=True
E 12 )
E 14 with Timer() as t:
E ---> 15 bivae.fit(train_set)
E 16 print("Took *** seconds for training.".format(t))
E
E File /azureml-envs/azureml_adf614c86c43311fb41235e[662](https://github.com/recommenders-team/recommenders/actions/runs/9745905406/job/26897451776#step:3:669)27b9b3/lib/python3.10/site-packages/cornac/models/bivaecf/recom_bivaecf.py:178, in BiVAECF.fit(self, train_set, val_set)
E 166 num_users = train_set.matrix.shape[0]
E 167 self.bivae = BiVAE(
E 168 k=self.k,
E 169 user_encoder_structure=[num_items] + self.encoder_structure,
E (...)
E 175 batch_size=self.batch_size,
E 176 ).to(self.device)
E --> 178 learn(
E 179 self.bivae,
E 180 train_set,
E 181 n_epochs=self.n_epochs,
E 182 batch_size=self.batch_size,
E 183 learn_rate=self.learning_rate,
E 184 beta_kl=self.beta_kl,
E 185 verbose=self.verbose,
E 186 device=self.device,
E 187 )
E 188 elif self.verbose:
E 189 print("%s is trained already (trainable = False)" % (self.name))
E
E File /azureml-envs/azureml_adf614c86c43311fb41235e66227b9b3/lib/python3.10/site-packages/cornac/models/bivaecf/bivae.py:201, in learn(bivae, train_set, n_epochs, batch_size, learn_rate, beta_kl, verbose, device, dtype)
E 199 for i_ids in train_set.item_iter(batch_size, shuffle=False):
E 200 i_batch = tx[i_ids, :]
E --> 201 i_batch = i_batch.A
E 202 i_batch = torch.tensor(i_batch, dtype=dtype, device=device)
E 204 # Reconstructed batch
E
E AttributeError: 'csc_matrix' object has no attribute 'A'
```
### In which platform does it happen?
<!--- Describe the platform where the issue is happening (use a list if needed) -->
<!--- For example: -->
<!--- * Azure Data Science Virtual Machine. -->
<!--- * Azure Databricks. -->
<!--- * Other platforms. -->
VM
### How do we replicate the issue?
<!--- Please be specific as possible (use a list if needed). -->
<!--- For example: -->
<!--- * Create a conda environment for pyspark -->
<!--- * Run unit test `test_sar_pyspark.py` with `pytest -m 'spark'` -->
<!--- * ... -->
https://github.com/recommenders-team/recommenders/actions/runs/9745905406/job/26897451776
### Expected behavior (i.e. solution)
<!--- For example: -->
<!--- * The tests for SAR PySpark should pass successfully. -->
### Other Comments
|
open
|
2024-07-07T10:34:09Z
|
2024-07-09T03:51:33Z
|
https://github.com/recommenders-team/recommenders/issues/2126
|
[
"bug"
] |
miguelgfierro
| 3
|
apachecn/ailearning
|
nlp
| 272
|
斗胆指一下注释中的一个小错误
|
https://github.com/apachecn/MachineLearning/blob/python-3.6/src/python/3.DecisionTree/DecisionTree.py
141行的注释 `# retDataSet = [data for data in dataSet for i, v in enumerate(data) if i == axis and v == value]` 这里的 `i == axis`是不是应该改为`i == index`?
|
closed
|
2018-01-28T02:43:41Z
|
2018-01-28T08:18:09Z
|
https://github.com/apachecn/ailearning/issues/272
|
[] |
ZhaoLizz
| 3
|
feature-engine/feature_engine
|
scikit-learn
| 810
|
`_is_categorical_and_is_datetime` with no default value
|
**Describe the bug**
`feature_engine.variable_handling._variable_type_checks._is_categorical_and_is_datetime` has variable `is_dt` has no default value
```bash
feature_engine/variable_handling/_variable_type_checks.py", line 52, in _is_categorical_and_is_datetime
return is_dt
^^^^^
UnboundLocalError: cannot access local variable 'is_dt' where it is not associated with a value
```
```
def _is_categorical_and_is_datetime(column: pd.Series) -> bool:
# check for datetime only if object cannot be cast as numeric because
# if it could pd.to_datetime would convert it to datetime regardless
if is_object(column):
is_dt = not _is_convertible_to_num(column) and _is_convertible_to_dt(column)
# check for datetime only if the type of the categories is not numeric
# because pd.to_datetime throws an error when it is an integer
elif isinstance(column.dtype, pd.CategoricalDtype):
is_dt = not _is_categories_num(column) and _is_convertible_to_dt(column)
return is_dt
```
should be turned into
```
def _is_categorical_and_is_datetime(column: pd.Series) -> bool:
# check for datetime only if object cannot be cast as numeric because
# if it could pd.to_datetime would convert it to datetime regardless
is_dt = False
if is_object(column):
is_dt = not _is_convertible_to_num(column) and _is_convertible_to_dt(column)
# check for datetime only if the type of the categories is not numeric
# because pd.to_datetime throws an error when it is an integer
elif isinstance(column.dtype, pd.CategoricalDtype):
is_dt = not _is_categories_num(column) and _is_convertible_to_dt(column)
return is_dt
```
|
closed
|
2024-09-03T07:56:38Z
|
2024-09-12T12:10:56Z
|
https://github.com/feature-engine/feature_engine/issues/810
|
[] |
jccalvojackson
| 2
|
ghtmtt/DataPlotly
|
plotly
| 6
|
Folder to save the plot
|
the `tmp` folder is the best solution at the moment.
**NOTE** make it cross platform!
|
closed
|
2017-05-08T15:29:46Z
|
2017-05-12T08:53:09Z
|
https://github.com/ghtmtt/DataPlotly/issues/6
|
[
"enhancement"
] |
ghtmtt
| 0
|
dadadel/pyment
|
numpy
| 63
|
Let NumpydocTools and GoogledocTools inherit from a common base class
|
As discussed in the abandoned (closed) PR #60, I open up another issue better suited to the changes I propose.
During modifications to the source code to fix issue #49, I realized that there was room for improvement in how the different docstring styles (Numpydoc and Googledoc) were handled. They had a lot of duplicate code and if they would have used a common base class, issue #49 should not have happened, as the GoogledocTools class already contained the solution.
My proposal is to introduce the common base class to prevent future issues arising because of duplicate logic in the now different "DocTools"
|
closed
|
2018-07-28T20:51:02Z
|
2018-08-01T12:21:07Z
|
https://github.com/dadadel/pyment/issues/63
|
[] |
wagnerpeer
| 1
|
nidhaloff/igel
|
automation
| 21
|
add cross validation support
|
### Description
Users should be able to use cross validation
|
closed
|
2020-09-28T20:08:50Z
|
2020-09-28T20:09:11Z
|
https://github.com/nidhaloff/igel/issues/21
|
[
"enhancement",
"feature"
] |
nidhaloff
| 1
|
AirtestProject/Airtest
|
automation
| 925
|
[BUG提交]
|
:bulb:**相关项目:** Airtest
**标题:** [BUG提交]
**AirtestIDE版本:**
**未使用本地Pyhton环境运行脚本**
**报错描述:**
无
**相关截图:**
无
**报错Log:**
无
**连接设备信息:**
无
**提供最小可复现此BUG的代码:**
```
无
```
|
closed
|
2021-06-25T08:21:14Z
|
2021-06-25T08:21:27Z
|
https://github.com/AirtestProject/Airtest/issues/925
|
[] |
NoneTypeCoder
| 0
|
SciTools/cartopy
|
matplotlib
| 1,684
|
Cartopy quiver plot issues - custom angles and updating quiver data
|
### Description
<!-- Please provide a general introduction to the issue/proposal. -->
I experience issues with the `angles` argument and the `set_UVC()` method for quiver plots when using the orthographic projection in Cartopy, see
see https://stackoverflow.com/questions/65021028/cartopy-quiver-plot-issues-custom-angles-and-updating-quiver-data
<!--
If you are reporting a bug, attach the *entire* traceback from Python.
If you are proposing an enhancement/new feature, provide links to related articles, reference examples, etc.
If you are asking a question, please ask on StackOverflow and use the cartopy tag. All cartopy
questions on StackOverflow can be found at https://stackoverflow.com/questions/tagged/cartopy
-->
#### Code to reproduce
```
```
#### Traceback
```
```
<details>
<summary>Full environment definition</summary>
<!-- fill in the following information as appropriate -->
### Operating system
### Cartopy version
### conda list
```
```
### pip list
```
```
</details>
|
open
|
2020-11-26T13:35:33Z
|
2021-02-20T13:22:58Z
|
https://github.com/SciTools/cartopy/issues/1684
|
[] |
silenceOfTheLambda
| 1
|
strawberry-graphql/strawberry
|
fastapi
| 3,381
|
Do the Lazy-Types support Union?
|
If I implement the following construct:
```
# graphql/file.py
if TYPE_CHECKING:
from graphql.status import StatusesResult
@strawberry.type
class File:
statuses: Annotated[
"StatusesResult", strawberry.lazy("graphql.status")
]
```
```
# graphql/status.py
@strawberry.type
class Status:
name: str
@strawberry.type
class Statuses:
statuses: list[Status]
StatusesResult = Annotated[
Union[Statuses, PermissionProblem],
strawberry.union("StatusesResult"),
]
```
I get this error when compiling:
```
TypeError: File fields cannot be resolved. Unexpected type 'typing.Annotated[typing.Union[graphql.status.Statuses, graphql.problem.PermissionProblem], <strawberry.union.StrawberryUnion object at 0x00000152A836DD10>]'
```
Have I implemented something incorrectly or is the support simply not yet available?
|
open
|
2024-02-14T10:46:02Z
|
2025-03-20T15:56:36Z
|
https://github.com/strawberry-graphql/strawberry/issues/3381
|
[] |
MaehMaeh
| 0
|
mljar/mljar-supervised
|
scikit-learn
| 133
|
Add feature selection step
|
Please add a feature selection step with the following steps:
1. Before the hill-climbing step add a random feature to the top-performing model.
2. Train the model with an additional random feature and compute permutation importance (for all features).
3. Drop all features which minimum importance is lower than the maximum importance of random feature across all trained learners.
4. Train a few models with a reduced number of features.
5. Perform standard hill-climbing step.
|
closed
|
2020-07-30T12:18:23Z
|
2020-07-31T10:48:14Z
|
https://github.com/mljar/mljar-supervised/issues/133
|
[
"enhancement"
] |
pplonski
| 0
|
iperov/DeepFaceLive
|
machine-learning
| 145
|
you should update yolov5 to yolor or yolov7 for more efficient face detection
|
you should update yolov5 to yolor or yolov7 for more efficient face detection https://github.com/WongKinYiu/yolor https://github.com/WongKinYiu/yolov7 use whatever version of either one is the most efficient and works the best.
|
closed
|
2023-04-09T08:05:39Z
|
2023-04-09T08:36:14Z
|
https://github.com/iperov/DeepFaceLive/issues/145
|
[] |
Cxsmo-ai
| 2
|
nok/sklearn-porter
|
scikit-learn
| 47
|
Export Matrix as Vector (SVM and maybe other Models)
|
Firstly, I wold like to thank the authors of the library, it is really useful.
Most of Java Algebra libraries are based on 1D primitive arrays (probably other languages too) instead of 2D (it is easy to map one to another and the algorithms in 1D are simpler to write). One option is to create a new 1D array and copy the data from the 2D, but it is not a desired approach. Then, I suggest that you provide a way to save the data as a 1D primitive array (more especially a 1D column array). I started doing this in a copy of the repository, but I guess you can do it in a future release.
I have an observation about the SVC template (I guess it should be in another place). When you save a model that has two classes, I guess the use of **starts** and **end** arrays are redundant, because **coefficients** is an ordered array (in the sense that all coefficients of the class zero are before any coefficient of the class one). It means you could change:
```java
...
if (this.clf.nClasses == 2) {
for (int i = 0; i < kernels.length; i++) {
kernels[i] = -kernels[i];
}
double decision = 0.;
for (int k = starts[1]; k < ends[1]; k++) {
decision += kernels[k] * this.clf.coefficients[0][k];
}
for (int k = starts[0]; k < ends[0]; k++) {
decision += kernels[k] * this.clf.coefficients[0][k];
}
decision += this.clf.intercepts[0];
if (decision > 0) {
return 0;
}
return 1;
}
...
```
to:
```java
...
if (this.clf.nClasses == 2) {
for (int i = 0; i < kernels.length; i++) {
kernels[i] = -kernels[i];
}
double decision = 0.;
for (int k = 0; k < clf.coefficients[0].length; k++) {
decision += kernels[k] * this.clf.coefficients[0][k];
}
decision += this.clf.intercepts[0];
if (decision > 0) {
return 0;
}
return 1;
}
...
```
I guess you could improve the case of more then two classes too, merging the structures **decisions**, **votes** and **amounts**.
Best Regards,
Charles
|
open
|
2019-02-09T19:02:15Z
|
2019-03-05T10:55:32Z
|
https://github.com/nok/sklearn-porter/issues/47
|
[
"enhancement"
] |
gobber
| 1
|
scikit-tda/kepler-mapper
|
data-visualization
| 103
|
Prediction for the missing segment of the blue sea star image
|
I really enjoyed reading this notebook: https://github.com/MLWave/kepler-mapper/tree/master/notebooks/self-guessing#references
I'm curious though: what do the predictions from a strong self-guesser for the "blue sea star" image at the start of the notebook look like?
|
closed
|
2018-07-10T18:23:52Z
|
2019-11-26T01:35:36Z
|
https://github.com/scikit-tda/kepler-mapper/issues/103
|
[] |
zachmayer
| 4
|
HumanSignal/labelImg
|
deep-learning
| 330
|
Difficult flag: missing instructions
|
hello.
Please add to readme some explanation regarding what the "difficult flag" is for and what difficulty means in terms of ML.
When and how exactly this flag meant to be used?
|
closed
|
2018-07-12T06:10:38Z
|
2018-07-15T08:21:18Z
|
https://github.com/HumanSignal/labelImg/issues/330
|
[] |
Insertfunnylogin
| 1
|
CPJKU/madmom
|
numpy
| 538
|
Start BeatTracker without Command line arguments
|
I have been trying to run the `BeatTracker` in my own class, so that I can can call a callback function whenever it detects a beat. However I cannot get it running without the command line argument "online"
Is there a way to get it running without command line arguments?
I have tried setting online to default in the parser, but nothing works.
I appreciate any help
|
open
|
2024-06-08T00:49:59Z
|
2024-06-10T18:43:02Z
|
https://github.com/CPJKU/madmom/issues/538
|
[] |
Jarris81
| 1
|
oegedijk/explainerdashboard
|
plotly
| 148
|
Adding new plotly graphs to create new components for custom dashboard
|
Hi
I'm trying to add a new graph to track predicted vs actual over time above the predicted vs actual component (for regression), like in the screenshot
But the graph shows as blank.
I created a function for the graph which works (index is dates):
```
def plotly_predicted_vs_actual_overtime(y, preds, target="" , units="", round=2, idxs=None, index_name="index"):
"""Generate graph showing predicted values from a regressor model vs actual
values over time.
Args:
y (np.ndarray): Actual values
preds (np.ndarray): Predicted values
target (str, optional): Label for target. Defaults to "".
units (str, optional): Units of target. Defaults to "".
round (int, optional): Rounding to apply to floats. Defaults to 2.
idxs (List[str], optional): list of identifiers for each observation. Defaults to None.
index_name (str): identifier for idxs. Defaults to "index".
Returns:
Plotly fig
"""
fig = go.Figure([
go.Scatter(
name='Predicted',
x=idxs,
y=preds,
mode='lines',
line=dict(color='rgb(0, 114, 178)'),
),
go.Scatter(
name='Upper Bound',
x=idxs,
y=preds+preds*0.1,
mode='lines',
marker=dict(color="#444"),
line=dict(width=0),
showlegend=False
),
go.Scatter(
name='Lower Bound',
x=idxs,
y=preds-preds*0.1,
marker=dict(color="#444"),
line=dict(width=0),
mode='lines',
fillcolor='rgba(68, 68, 68, 0.3)',
fill='tonexty',
showlegend=False
),
go.Scatter(
name='Actual',
x=idxs,
y=y,
mode='lines',
line=dict(color='rgb(213, 94, 0)'),
),
])
fig.update_layout(
yaxis_title=f"{target}" + (f" ({units})" if units else ""),
xaxis_title="date",
title="Predicted vs Actual "+ f"{target}"+ " over time",
hovermode="x"
)
fig.update_xaxes(
rangeslider_visible=True,
rangeselector=dict(
buttons=list([
dict(count=1, label="1m", step="month", stepmode="backward"),
dict(count=6, label="6m", step="month", stepmode="backward"),
dict(count=1, label="YTD", step="year", stepmode="todate"),
dict(count=1, label="1y", step="year", stepmode="backward"),
dict(step="all")
])
)
)
fig.update_traces(hovertemplate='%{y:.2f}'+units)
return fig
```
I saw that another function is created under RegressionExplainer like:
https://github.com/oegedijk/explainerdashboard/blob/29d25ba4ac867d32150933f00bcdfe95d0309894/explainerdashboard/explainers.py#L3346
Do I need to insert a "plot" function under this to solve this problem? From what I can see the way it works is plotly function -> plot function (under RegressionExplainer) -> Component Class
Have I missed anything to show the graph in my screenshot?
|
closed
|
2021-09-22T21:43:58Z
|
2021-12-23T19:13:44Z
|
https://github.com/oegedijk/explainerdashboard/issues/148
|
[] |
yean8mun
| 1
|
flairNLP/fundus
|
web-scraping
| 1
|
Move current repo
|
- [ ] Get current [repo](https://github.com/MaxDall/CCNewsCrawler) in a movable state (commit all changes, etc...) -
- [ ] Move repo
|
closed
|
2022-10-28T16:14:48Z
|
2022-10-31T13:41:18Z
|
https://github.com/flairNLP/fundus/issues/1
|
[] |
MaxDall
| 4
|
FactoryBoy/factory_boy
|
django
| 160
|
Model factory should load model as late as possible
|
Django uses string notation in several places (e.g. `ForeignKey`) to be able to reference a model during the app startup phase. Factory boy recently added that syntax, but unfortunately due to the way it's implemented, the original purpose of lazy loading the model is defeated.
The model class is loaded in the factory's Meta class `__new__` method. That means at import time, not when the factory is first instantiated. The model class is [loaded](https://github.com/rbarrois/factory_boy/blob/master/factory/django.py#L93) via Django's `get_model` method. That means using the string syntax is actually _worse_ than directly declaring the model, because at least on Django 1.7, `get_model` fails loudly if the entire app cache isn't populated yet, while passing the model lets Django deal with the population of the app cache.
This leads to all kinds of app startup problems when e.g. collecting tests for a test suite, because it suddenly requires the app cache to be fully initialised. But while directly referencing the model as a workaround is possible, it can still lead to tricky situations.
One way to fix it might be to only load the model class when the factory itself is initialised.
|
open
|
2014-08-28T19:45:35Z
|
2015-01-30T16:21:20Z
|
https://github.com/FactoryBoy/factory_boy/issues/160
|
[
"Bug"
] |
maiksprenger
| 6
|
Esri/arcgis-python-api
|
jupyter
| 2,149
|
Unable to get arcgis.mapping to work
|
**Describe the bug**
Unable to use the arcgis mapping module
error:
```python
Import Error: Cannot import WebMap from arcgis.mapping
```
**Screenshots**
<img width="1394" alt="Screenshot 2024-10-29 at 2 30 09 PM" src="https://github.com/user-attachments/assets/4174f044-8c1c-4d0d-8c35-5a81e3f2859d">
**Platform (please complete the following information):**
- OS: [Docker] https://github.com/Esri/arcgis-python-api/pkgs/container/arcgis-python-api-notebook/283262919?tag=latest
- Browser [Chrome]
- Python API Version [`2.4.0`]
**Additional context**
I used the docker availble on the repo, the same is reproducible if using it in a conda environment or on Google Colab with `!pip install arcgis`
|
closed
|
2024-10-29T19:30:27Z
|
2024-10-30T16:48:08Z
|
https://github.com/Esri/arcgis-python-api/issues/2149
|
[
"As-Designed"
] |
gv2325
| 6
|
ivy-llc/ivy
|
pytorch
| 28,619
|
Fix Frontend Failing Test: numpy - search.paddle.argsort
|
To-do List: https://github.com/unifyai/ivy/issues/27497
|
closed
|
2024-03-17T14:15:22Z
|
2024-03-25T12:44:28Z
|
https://github.com/ivy-llc/ivy/issues/28619
|
[
"Sub Task"
] |
ZJay07
| 0
|
encode/apistar
|
api
| 294
|
Test cli arguments
|
To run `pytest` over the project layout below, it would be needed the argument `--pyargs project`. Since `apistar test` run pytest, would be great to pass its args/kwargs e.g. `apistar test --pyargs project`
```
project/
__init__.py
app.py
view.py
test/
__init__.py
test_app.py
test_view.py
...
```
More on: https://docs.pytest.org/en/latest/goodpractices.html#test-discovery
EDIT:
Obviously you have a lot of thoughts on this. Since the project I'm working on right now is using apistar, I saw a great opportunity to help the project.
|
closed
|
2017-09-22T03:28:29Z
|
2018-03-28T21:22:39Z
|
https://github.com/encode/apistar/issues/294
|
[] |
rougeth
| 1
|
albumentations-team/albumentations
|
machine-learning
| 2,402
|
[New feature] Add apply_to_images to FancyPCA
|
open
|
2025-03-11T01:02:29Z
|
2025-03-11T01:02:35Z
|
https://github.com/albumentations-team/albumentations/issues/2402
|
[
"enhancement",
"good first issue"
] |
ternaus
| 0
|
|
indico/indico
|
sqlalchemy
| 5,956
|
Room booking: Rich-text room descriptions / multiple room pictures
|
**Is your feature request related to a problem? Please describe.**
For some rooms, more than one picture is needed to communicate everything — for example, the usual layout of tables and chairs (or, if there are several layouts, a list of them), special equipment etc.
The rooms currently only allow for a single picture and a plain-text comment.
**Describe the solution you'd like**
It would be nice to have a full-fledged description field, similarly to what is possible for the "Description" of events.
This would allow to add multiple pictures, if needed, and more elaborate descriptions (for things which don't fit into equipment, e.g. the geographic direction of windows, different chair / table layouts etc.).
**Describe alternatives you've considered**
Linking to a different page for each room adding more details — but this adds the additional complexity of having a different system with similar access permissions and reachability.
**Additional context**
The main request here was to add a way to show different possible chair / table arrangements, e.g. in a top view. Of course, a photo of the room should still be possible.
So technically, another possible solution instead of adding a rich formatting field would be to add a way to add multiple pictures for a given room only.
|
open
|
2023-09-27T19:12:27Z
|
2023-09-27T22:12:56Z
|
https://github.com/indico/indico/issues/5956
|
[
"enhancement"
] |
olifre
| 2
|
Miserlou/Zappa
|
django
| 1,921
|
-bash: zappa: command not found
|
<!--- Provide a general summary of the issue in the Title above -->
## Context
i solved my issue by uninstalling pip and python and re setting up the environment and installation ect
|
closed
|
2019-08-20T20:41:42Z
|
2022-11-20T11:54:58Z
|
https://github.com/Miserlou/Zappa/issues/1921
|
[] |
3lonious
| 1
|
SYSTRAN/faster-whisper
|
deep-learning
| 674
|
ValueError: invalid literal for int() with base 10: ''
|
faster-whisper: 0.10.0
```
bash[610]: Traceback (most recent call last):
bash[610]: File "../app.py", line 17, in <module>
bash[610]: from faster_whisper import WhisperModel
bash[610]: File "/opt/conda/envs/pytorch/lib/python3.9/site-packages/faster_whisper/__init__.py", line 2, in <module>
bash[610]: from faster_whisper.transcribe import WhisperModel
bash[610]: File "/opt/conda/envs/pytorch/lib/python3.9/site-packages/faster_whisper/transcribe.py", line 10, in <module>
bash[610]: import ctranslate2
bash[610]: File "/opt/conda/envs/pytorch/lib/python3.9/site-packages/ctranslate2/__init__.py", line 53, in <module>
bash[610]: from ctranslate2 import converters, models, specs
bash[610]: File "/opt/conda/envs/pytorch/lib/python3.9/site-packages/ctranslate2/converters/__init__.py", line 8, in <module>
bash[610]: from ctranslate2.converters.transformers import TransformersConverter
bash[610]: File "/opt/conda/envs/pytorch/lib/python3.9/site-packages/ctranslate2/converters/transformers.py", line 14, in <module>
bash[610]: import transformers
bash[610]: File "/opt/conda/envs/pytorch/lib/python3.9/site-packages/transformers/__init__.py", line 30, in <module>
bash[610]: from . import dependency_versions_check
bash[610]: File "/opt/conda/envs/pytorch/lib/python3.9/site-packages/transformers/dependency_versions_check.py", line 17, in <module>
bash[610]: from .utils.versions import require_version, require_version_core
bash[610]: File "/opt/conda/envs/pytorch/lib/python3.9/site-packages/transformers/utils/__init__.py", line 60, in <module>
bash[610]: from .hub import (
bash[610]: File "/opt/conda/envs/pytorch/lib/python3.9/site-packages/transformers/utils/hub.py", line 1087, in <module>
bash[610]: cache_version = int(f.read())
bash[610]: ValueError: invalid literal for int() with base 10: ''
```
|
open
|
2024-02-08T02:58:52Z
|
2024-02-08T02:58:52Z
|
https://github.com/SYSTRAN/faster-whisper/issues/674
|
[] |
tree1891
| 0
|
iperov/DeepFaceLab
|
machine-learning
| 893
|
Please add 3 pass in face detection
|
Please add 3 pass in face detection just like in DFL 1.0 cause too many false face detections
|
open
|
2020-09-15T01:23:11Z
|
2023-06-08T21:16:31Z
|
https://github.com/iperov/DeepFaceLab/issues/893
|
[] |
justinjohn0306
| 1
|
horovod/horovod
|
machine-learning
| 3,493
|
Training loop now working very slow
|
Hello !
I am sorry but after long struggling installing the horovod now I am facing new strange issue. My code run now very slowly comparing to before.
Also using nccl configuration I am getting this warning
```
[0]<stderr>:libibverbs: Warning: couldn't load driver 'librxe-rdmav34.so': librxe-rdmav34.so: cannot open shared object file: No such file or directory
[0]<stderr>:libibverbs: Warning: couldn't load driver 'libmlx4-rdmav34.so': libmlx4-rdmav34.so: cannot open shared object file: No such file or directory
[4]<stderr>:libibverbs: Warning: couldn't load driver 'librxe-rdmav34.so': librxe-rdmav34.so: cannot open shared object file: No such file or directory
[4]<stderr>:libibverbs: Warning: couldn't load driver 'libmlx4-rdmav34.so': libmlx4-rdmav34.so: cannot open shared object file: No such file or directory
[2]<stderr>:libibverbs: Warning: couldn't load driver 'librxe-rdmav34.so': librxe-rdmav34.so: cannot open shared object file: No such file or directory
[2]<stderr>:libibverbs: Warning: couldn't load driver 'libmlx4-rdmav34.so': libmlx4-rdmav34.so: cannot open shared object file: No such file or directory
[5]<stderr>:libibverbs: Warning: couldn't load driver 'librxe-rdmav34.so': librxe-rdmav34.so: cannot open shared object file: No such file or directory
[7]<stderr>:libibverbs: Warning: couldn't load driver 'librxe-rdmav34.so': librxe-rdmav34.so: cannot open shared object file: No such file or directory
[6]<stderr>:libibverbs: Warning: couldn't load driver 'librxe-rdmav34.so': librxe-rdmav34.so: cannot open shared object file: No such file or directory
[1]<stderr>:libibverbs: Warning: couldn't load driver 'librxe-rdmav34.so': librxe-rdmav34.so: cannot open shared object file: No such file or directory
[5]<stderr>:libibverbs: Warning: couldn't load driver 'libmlx4-rdmav34.so': libmlx4-rdmav34.so: cannot open shared object file: No such file or directory
[7]<stderr>:libibverbs: Warning: couldn't load driver 'libmlx4-rdmav34.so': libmlx4-rdmav34.so: cannot open shared object file: No such file or directory
[6]<stderr>:libibverbs: Warning: couldn't load driver 'libmlx4-rdmav34.so': libmlx4-rdmav34.so: cannot open shared object file: No such file or directory
[1]<stderr>:libibverbs: Warning: couldn't load driver 'libmlx4-rdmav34.so': libmlx4-rdmav34.so: cannot open shared object file: No such file or directory
[3]<stderr>:libibverbs: Warning: couldn't load driver 'librxe-rdmav34.so': librxe-rdmav34.so: cannot open shared object file: No such file or directory
[3]<stderr>:libibverbs: Warning: couldn't load driver 'libmlx4-rdmav34.so': libmlx4-rdmav34.so: cannot open shared object file: No such file or directory
```
which disappears when excluding nccl from installation.
Details of horovod installation:
Env was installed following these [steps](https://github.com/Arij-Aladel/T5-Tasks/blob/main/Readme.md#install-horovod)
My question why training using horovod became so so slow? is it related to the server? to the env configuration?
FYI; it was working quickly with the same env setup.
I am using very powerful server A100 with 40 GB memory and 8 gpus. The training time increased from 23 sec in average into 5 minutes in average for each batch. I know with such details you can not draw full image of the issue so please tell me which details help to understand the reasons of this issue so I can provide.
|
closed
|
2022-03-25T09:03:50Z
|
2022-06-10T18:24:05Z
|
https://github.com/horovod/horovod/issues/3493
|
[
"wontfix"
] |
Arij-Aladel
| 1
|
mouredev/Hello-Python
|
fastapi
| 79
|
Python
|
open
|
2024-10-02T16:24:12Z
|
2024-10-02T16:24:12Z
|
https://github.com/mouredev/Hello-Python/issues/79
|
[] |
Ivan-Reartes
| 0
|
|
plotly/dash-table
|
dash
| 313
|
Ability to export data as excel or csv
|
- For Excel files, only XLSX (not XLS) will be supported
- Only the data will be exported, formatting will not be exported
- The export will include the data in the current view. For example, if columns are hidden, sorted, or filtered, then the exported file will display the current view.
- Export will not protect users from “CSV Injection” attacks (https://www.owasp.org/index.php/CSV_Injection)
Exporting Excel files may require a large 3rd party open source library. This library is too large to include in the default version of dash-table, so we will need to engineer dynamic JavaScript module loading as part of this requirement.
We will consider the UI for this feature in a separate issue (we need to design a UI needs encompasses all of the new features that we're adding).
|
closed
|
2018-12-19T22:02:52Z
|
2019-08-08T20:28:38Z
|
https://github.com/plotly/dash-table/issues/313
|
[
"dash-type-enhancement",
"dash-meta-sponsored",
"size: 8"
] |
chriddyp
| 8
|
sktime/sktime
|
data-science
| 8,021
|
[ENH] Inconsistent behaviour in keeping track of index name between pandas.Series and pandas.DataFrame
|
`sktime` is not keeping index name information consistently. As a user, i expect it to be kept track either always or never.
```pycon
>>>
>>> import pandas
>>> from sktime.forecasting.ets import AutoETS
>>>
>>> pandas_df = pandas.DataFrame({"date": pandas.date_range(start="2001-01-01", end="2001-01-04", freq="D"), "y1": [1, 2, 3, 4], "y2": [11, 12, 13, 14], "y3": [21, 22, 23, 24]})
>>> sktime_df = pandas_df.set_index("date")
>>>
>>> series_example = sktime_df["y3"]
>>> dataframe_example = sktime_df[["y1", "y2"]]
>>>
>>> series_forecaster = AutoETS()
>>> _ = series_forecaster.fit(series_example)
/home/anirban/conda-environments/sktime/lib/python3.10/site-packages/statsmodels/tsa/base/tsa_model.py:473: ValueWarning: No frequency information was provided, so inferred frequency D will be used.
self._init_dates(dates, freq)
>>> series_forecaster.predict(fh=[1]) # index name "date" is lost
2001-01-05 23.9999
Name: y3, dtype: float64
>>>
>>> dataframe_forecaster = AutoETS()
>>> _ = dataframe_forecaster.fit(dataframe_example)
>>> dataframe_forecaster.predict(fh=[1]) # index name "date" is kept
y1 y2
date
2001-01-05 3.9999 13.9999
>>>
```
|
open
|
2025-03-22T10:06:51Z
|
2025-03-23T23:04:39Z
|
https://github.com/sktime/sktime/issues/8021
|
[
"module:datatypes",
"enhancement"
] |
yarnabrina
| 7
|
tflearn/tflearn
|
tensorflow
| 1,140
|
List index out of range for FashionMNIST tutorial.
|
I've just started learning a bit about Tensorflow, and I tried out the Fashion MNIST tutorial. However, I keep getting a list index out of range error. I've copied the code from the official tutorial and also tried using tf.reset_default_graph() as suggested in some other posts, but neither have worked.
Here is the notebook with the error:
https://github.com/fsiraj/Tensorflow-Tutorials/blob/master/Fashion%20MNIST.ipynb
Apologies if I've posted this in the wrong repo, I'm not sure if this issue should be posted here.
|
open
|
2019-11-05T22:17:14Z
|
2019-11-05T22:26:23Z
|
https://github.com/tflearn/tflearn/issues/1140
|
[] |
fsiraj
| 0
|
public-apis/public-apis
|
api
| 3,321
|
Apis
|
closed
|
2022-10-06T16:22:16Z
|
2022-10-14T08:01:37Z
|
https://github.com/public-apis/public-apis/issues/3321
|
[] |
S1mon009
| 0
|
|
harry0703/MoneyPrinterTurbo
|
automation
| 489
|
视频生成失败
|
2024-09-03 16:04:29.288 | INFO | __main__:<module>:783 - 开始生成视频
2024-09-03 16:04:29.308 | INFO | __main__:<module>:784 - {
"video_subject": "父母如何教导孩子学会拒绝",
"video_script": "",
"video_terms": "",
"video_aspect": "16:9",
"video_concat_mode": "random",
"video_clip_duration": 3,
"video_count": 1,
"video_source": "pixabay",
"video_materials": null,
"video_language": "zh-CN",
"voice_name": "zh-CN-XiaoxiaoNeural-Female",
"voice_volume": 1.0,
"voice_rate": 1.0,
"bgm_type": "random",
"bgm_file": "",
"bgm_volume": 0.2,
"subtitle_enabled": true,
"subtitle_position": "bottom",
"custom_position": 70.0,
"font_name": "MicrosoftYaHeiNormal.ttc",
"text_fore_color": "#e4128f",
"text_background_color": "transparent",
"font_size": 60,
"stroke_color": "#000000",
"stroke_width": 1.5,
"n_threads": 2,
"paragraph_number": 1
}
2024-09-03 16:04:29.324 | INFO | app.services.task:start:210 - start task: ede388e9-37c0-4f86-a79f-0bc31d76d54e, stop_at: video
2024-09-03 16:04:29.327 | INFO | app.services.task:generate_script:18 -
## generating video script
2024-09-03 16:04:29.330 | INFO | app.services.llm:generate_script:282 - subject: 父母如何教导孩子学会拒绝
2024-09-03 16:04:29.332 | INFO | app.services.llm:_generate_response:18 - llm provider: moonshot
2024-09-03 16:04:32.142 | ERROR | app.services.llm:generate_script:318 - failed to generate script: Error code: 401 - {'error': {'message': 'Invalid Authentication', 'type': 'invalid_authentication_error'}}
2024-09-03 16:04:32.144 | WARNING | app.services.llm:generate_script:321 - failed to generate video script, trying again... 1
2024-09-03 16:04:32.146 | INFO | app.services.llm:_generate_response:18 - llm provider: moonshot
2024-09-03 16:04:33.646 | ERROR | app.services.llm:generate_script:318 - failed to generate script: Error code: 401 - {'error': {'message': 'Incorrect API key provided', 'type': 'incorrect_api_key_error'}}
2024-09-03 16:04:33.648 | WARNING | app.services.llm:generate_script:321 - failed to generate video script, trying again... 2
2024-09-03 16:04:33.651 | INFO | app.services.llm:_generate_response:18 - llm provider: moonshot
2024-09-03 16:04:36.265 | ERROR | app.services.llm:generate_script:318 - failed to generate script: Error code: 401 - {'error': {'message': 'Invalid Authentication', 'type': 'invalid_authentication_error'}}
2024-09-03 16:04:36.267 | WARNING | app.services.llm:generate_script:321 - failed to generate video script, trying again... 3
2024-09-03 16:04:36.268 | INFO | app.services.llm:_generate_response:18 - llm provider: moonshot
2024-09-03 16:04:38.717 | ERROR | app.services.llm:generate_script:318 - failed to generate script: Error code: 401 - {'error': {'message': 'Invalid Authentication', 'type': 'invalid_authentication_error'}}
2024-09-03 16:04:38.718 | WARNING | app.services.llm:generate_script:321 - failed to generate video script, trying again... 4
2024-09-03 16:04:38.719 | INFO | app.services.llm:_generate_response:18 - llm provider: moonshot
2024-09-03 16:04:41.196 | ERROR | app.services.llm:generate_script:318 - failed to generate script: Error code: 401 - {'error': {'message': 'Invalid Authentication', 'type': 'invalid_authentication_error'}}
2024-09-03 16:04:41.198 | WARNING | app.services.llm:generate_script:321 - failed to generate video script, trying again... 5
2024-09-03 16:04:41.199 | SUCCESS | app.services.llm:generate_script:323 - completed:
2024-09-03 16:04:41.200 | ERROR | app.services.task:generate_script:31 - failed to generate video script.
2024-09-03 16:04:41.201 | ERROR | __main__:<module>:790 - 视频生成失败
|
open
|
2024-09-03T08:07:05Z
|
2024-09-23T07:03:05Z
|
https://github.com/harry0703/MoneyPrinterTurbo/issues/489
|
[] |
Jiang8002000
| 2
|
jstrieb/github-stats
|
asyncio
| 62
|
display by line number
|
Is is possible to display languages used by line number?
|
closed
|
2022-01-20T19:59:42Z
|
2022-03-04T14:15:29Z
|
https://github.com/jstrieb/github-stats/issues/62
|
[] |
hhaootian
| 2
|
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.