repo_name
stringlengths 9
75
| topic
stringclasses 30
values | issue_number
int64 1
203k
| title
stringlengths 1
976
| body
stringlengths 0
254k
| state
stringclasses 2
values | created_at
stringlengths 20
20
| updated_at
stringlengths 20
20
| url
stringlengths 38
105
| labels
listlengths 0
9
| user_login
stringlengths 1
39
| comments_count
int64 0
452
|
|---|---|---|---|---|---|---|---|---|---|---|---|
SciTools/cartopy
|
matplotlib
| 2,482
|
indicate_inset_zoom not working on some projections
|
### Description
On most non-standard projections (like `TransverseMercator`) the connectors to an inset axis are not drawn correctly.
#### Code to reproduce
```python
map_proj = ccrs.TransverseMercator()
fig, ax = plt.subplots(
figsize=(15, 15),
subplot_kw={
"projection": map_proj
},
)
ax.set_extent([-50, 15, 50, 70], crs=ccrs.PlateCarree())
ax.add_feature(cfeature.OCEAN.with_scale('50m'), facecolor="lightsteelblue", zorder=1)
ax.coastlines(linewidth=0.2, zorder=2, resolution='50m')
# Add inset
inset_ax = inset_axes(
ax,
width="30%", # Width as a percentage of the parent axes
height="30%", # Height as a percentage of the parent axes
loc='lower left', # Location of the inset
bbox_to_anchor=(0, 0, 1, 1),
bbox_transform=ax.transAxes,
axes_class=GeoAxes,
axes_kwargs=dict(projection=map_proj)
)
inset_extent = [-43, -39, 63, 66]
inset_ax.set_extent(inset_extent, crs=ccrs.PlateCarree())
# Add features to inset
inset_ax.add_feature(cfeature.OCEAN, facecolor="lightsteelblue", zorder=1)
inset_ax.coastlines(linewidth=0.5, zorder=2, resolution='50m')
# Add box around location of inset map on the main map
x = [inset_extent[0], inset_extent[1], inset_extent[1], inset_extent[0], inset_extent[0]]
y = [inset_extent[2], inset_extent[2], inset_extent[3], inset_extent[3], inset_extent[2]]
ax.plot(x, y, color='k', alpha=0.5, transform=ccrs.PlateCarree())
# Draw lines between inset map and box on main map
rect, connectors = ax.indicate_inset_zoom(inset_ax, edgecolor="black", alpha=0.5, transform=ax.transAxes)
```
produces this

However, for some projections like `Mercator` or `PlateCarree`, it works

<details>
<summary>Full environment definition</summary>
### Cartopy version
0.24.0
### conda list
```
# Name Version Build Channel
aemet-opendata 0.5.4 pypi_0 pypi
aenum 3.1.15 pyhd8ed1ab_0 conda-forge
affine 2.4.0 pyhd8ed1ab_0 conda-forge
aiohttp 3.9.5 py312h41838bb_0 conda-forge
aiosignal 1.3.1 pyhd8ed1ab_0 conda-forge
annotated-types 0.7.0 pyhd8ed1ab_0 conda-forge
anyio 4.6.0 pyhd8ed1ab_1 conda-forge
appdirs 1.4.4 pyh9f0ad1d_0 conda-forge
appnope 0.1.4 pyhd8ed1ab_0 conda-forge
argon2-cffi 23.1.0 pyhd8ed1ab_0 conda-forge
argon2-cffi-bindings 21.2.0 py312hb553811_5 conda-forge
arrow 1.3.0 pyhd8ed1ab_0 conda-forge
asciitree 0.3.3 py_2 conda-forge
asttokens 2.4.1 pyhd8ed1ab_0 conda-forge
async-lru 2.0.4 pyhd8ed1ab_0 conda-forge
attrs 24.2.0 pyh71513ae_0 conda-forge
aws-c-auth 0.7.30 h0e83244_0 conda-forge
aws-c-cal 0.7.4 h8128ea2_1 conda-forge
aws-c-common 0.9.28 h00291cd_0 conda-forge
aws-c-compression 0.2.19 h8128ea2_1 conda-forge
aws-c-event-stream 0.4.3 hcd1ed9e_2 conda-forge
aws-c-http 0.8.9 h2f86973_0 conda-forge
aws-c-io 0.14.18 hf9a0f1c_10 conda-forge
aws-c-mqtt 0.10.5 h3e3652f_0 conda-forge
aws-c-s3 0.6.5 hf761692_5 conda-forge
aws-c-sdkutils 0.1.19 h8128ea2_3 conda-forge
aws-checksums 0.1.20 h8128ea2_0 conda-forge
aws-crt-cpp 0.28.2 heb037f6_6 conda-forge
aws-sdk-cpp 1.11.379 hd82b0e1_10 conda-forge
azure-core-cpp 1.13.0 hf8dbe3c_0 conda-forge
azure-identity-cpp 1.8.0 h60298e3_2 conda-forge
azure-storage-blobs-cpp 12.12.0 h646f05d_0 conda-forge
azure-storage-common-cpp 12.7.0 hf91904f_1 conda-forge
azure-storage-files-datalake-cpp 12.11.0 h14965f0_1 conda-forge
babel 2.14.0 pyhd8ed1ab_0 conda-forge
backports-datetime-fromisoformat 2.0.2 py312hb401068_0 conda-forge
basemap 1.4.1 np126py312hb3450bc_0 conda-forge
basemap-data 1.3.2 pyhd8ed1ab_3 conda-forge
basemap-data-hires 1.3.2 pyhd8ed1ab_3 conda-forge
beautifulsoup4 4.12.3 pyha770c72_0 conda-forge
bleach 6.1.0 pyhd8ed1ab_0 conda-forge
blinker 1.8.2 pyhd8ed1ab_0 conda-forge
blosc 1.21.6 h7d75f6d_0 conda-forge
bokeh 3.5.2 pyhd8ed1ab_0 conda-forge
boltons 24.0.0 pyhd8ed1ab_0 conda-forge
boto3 1.35.34 pyhd8ed1ab_0 conda-forge
botocore 1.35.34 pyge310_1234567_0 conda-forge
bottleneck 1.4.0 py312h3a11e2b_2 conda-forge
branca 0.7.2 pyhd8ed1ab_0 conda-forge
brotli 1.1.0 h00291cd_2 conda-forge
brotli-bin 1.1.0 h00291cd_2 conda-forge
brotli-python 1.1.0 py312h5861a67_2 conda-forge
bzip2 1.0.8 hfdf4475_7 conda-forge
c-ares 1.33.1 h44e7173_0 conda-forge
ca-certificates 2024.8.30 h8857fd0_0 conda-forge
cached-property 1.5.2 hd8ed1ab_1 conda-forge
cached_property 1.5.2 pyha770c72_1 conda-forge
cachelib 0.9.0 pyhd8ed1ab_0 conda-forge
cachetools 5.5.0 pyhd8ed1ab_0 conda-forge
cachier 3.0.1 pyhd8ed1ab_0 conda-forge
cairo 1.18.0 h37bd5c4_3 conda-forge
cartopy 0.24.0 py312h98e817e_0 conda-forge
cctools_osx-64 973.0.1 habff3f6_15 conda-forge
cdo 2.4.1 h70e7d24_1 conda-forge
certifi 2024.8.30 pyhd8ed1ab_0 conda-forge
cf_xarray 0.9.5 pyhd8ed1ab_1 conda-forge
cffi 1.17.1 py312hf857d28_0 conda-forge
cfgrib 0.9.14.1 pyhd8ed1ab_0 conda-forge
cfitsio 4.4.1 ha105788_0 conda-forge
cftime 1.6.4 py312h3a11e2b_1 conda-forge
charset-normalizer 3.3.2 pyhd8ed1ab_0 conda-forge
clang 15.0.7 hdae98eb_5 conda-forge
clang-15 15.0.7 default_h7151d67_5 conda-forge
clang_impl_osx-64 15.0.7 h03d6864_8 conda-forge
clang_osx-64 15.0.7 hb91bd55_8 conda-forge
clangxx 15.0.7 default_h7151d67_5 conda-forge
clangxx_impl_osx-64 15.0.7 h2133e9c_8 conda-forge
clangxx_osx-64 15.0.7 hb91bd55_8 conda-forge
click 8.1.7 unix_pyh707e725_0 conda-forge
click-params 0.5.0 pyhd8ed1ab_0 conda-forge
click-plugins 1.1.1 py_0 conda-forge
cligj 0.7.2 pyhd8ed1ab_1 conda-forge
cloudpickle 3.0.0 pyhd8ed1ab_0 conda-forge
cloup 3.0.5 pyhd8ed1ab_0 conda-forge
cmdstan 2.33.1 ha749d2a_0 conda-forge
cmdstanpy 1.2.4 pyhd8ed1ab_0 conda-forge
cmweather 0.3.2 pyhd8ed1ab_0 conda-forge
colorama 0.4.6 pyhd8ed1ab_0 conda-forge
colorcet 3.1.0 pyhd8ed1ab_0 conda-forge
comm 0.2.2 pyhd8ed1ab_0 conda-forge
compiler-rt 15.0.7 ha38d28d_2 conda-forge
compiler-rt_osx-64 15.0.7 ha38d28d_2 conda-forge
configobj 5.0.9 pyhd8ed1ab_0 conda-forge
contourpy 1.3.0 py312hc5c4d5f_2 conda-forge
convertdate 2.4.0 pyhd8ed1ab_0 conda-forge
copernicusmarine 1.3.3 pyhd8ed1ab_0 conda-forge
cycler 0.12.1 pyhd8ed1ab_0 conda-forge
cyrus-sasl 2.1.27 hf9bab2b_7 conda-forge
cytoolz 1.0.0 py312hb553811_0 conda-forge
dash 2.18.1 pyhd8ed1ab_0 conda-forge
dash-bootstrap-components 1.6.0 pyhd8ed1ab_0 conda-forge
dash-iconify 0.1.2 pyhd8ed1ab_0 conda-forge
dash-leaflet 1.0.15 pyhd8ed1ab_0 conda-forge
dash-mantine-components 0.14.4 pyhd8ed1ab_0 conda-forge
dask 2024.9.1 pyhd8ed1ab_0 conda-forge
dask-core 2024.9.1 pyhd8ed1ab_0 conda-forge
dask-expr 1.1.15 pyhd8ed1ab_0 conda-forge
dataclasses 0.8 pyhc8e2a94_3 conda-forge
debugpy 1.8.6 py312h5861a67_0 conda-forge
decorator 5.1.1 pyhd8ed1ab_0 conda-forge
defusedxml 0.7.1 pyhd8ed1ab_0 conda-forge
deprecated 1.2.14 pyh1a96a4e_0 conda-forge
deprecation 2.1.0 pyh9f0ad1d_0 conda-forge
dill 0.3.9 pyhd8ed1ab_0 conda-forge
diskcache 5.6.3 pyhd8ed1ab_0 conda-forge
distributed 2024.9.1 pyhd8ed1ab_0 conda-forge
distro 1.9.0 pyhd8ed1ab_0 conda-forge
dnspython 2.7.0 pyhff2d567_0 conda-forge
docopt 0.6.2 py_1 conda-forge
docopt-ng 0.9.0 pyhd8ed1ab_0 conda-forge
donfig 0.8.1.post1 pyhd8ed1ab_0 conda-forge
eccodes 2.38.0 he0f85d2_0 conda-forge
ecmwf-opendata 0.3.10 pyhd8ed1ab_0 conda-forge
entrypoints 0.4 pyhd8ed1ab_0 conda-forge
environs 11.0.0 pyhd8ed1ab_1 conda-forge
ephem 4.1.5 py312hb553811_2 conda-forge
eumdac 2.2.3 pyhd8ed1ab_0 conda-forge
eval-type-backport 0.2.0 pyhd8ed1ab_0 conda-forge
eval_type_backport 0.2.0 pyha770c72_0 conda-forge
exceptiongroup 1.2.2 pyhd8ed1ab_0 conda-forge
executing 2.1.0 pyhd8ed1ab_0 conda-forge
expat 2.6.3 hac325c4_0 conda-forge
fasteners 0.17.3 pyhd8ed1ab_0 conda-forge
fftw 3.3.10 nompi_h292e606_110 conda-forge
filelock 3.16.1 pyhd8ed1ab_0 conda-forge
findlibs 0.0.5 pyhd8ed1ab_0 conda-forge
fiona 1.9.6 py312hfc836c0_3 conda-forge
flask 3.0.3 pyhd8ed1ab_0 conda-forge
flask-caching 2.1.0 pyhd8ed1ab_0 conda-forge
flexcache 0.3 pyhd8ed1ab_0 conda-forge
flexparser 0.3.1 pyhd8ed1ab_0 conda-forge
fmt 11.0.2 h3c5361c_0 conda-forge
folium 0.17.0 pyhd8ed1ab_0 conda-forge
font-ttf-dejavu-sans-mono 2.37 hab24e00_0 conda-forge
font-ttf-inconsolata 3.000 h77eed37_0 conda-forge
font-ttf-source-code-pro 2.038 h77eed37_0 conda-forge
font-ttf-ubuntu 0.83 h77eed37_3 conda-forge
fontconfig 2.14.2 h5bb23bf_0 conda-forge
fonts-conda-ecosystem 1 0 conda-forge
fonts-conda-forge 1 0 conda-forge
fonttools 4.54.1 py312hb553811_0 conda-forge
fqdn 1.5.1 pyhd8ed1ab_0 conda-forge
freetype 2.12.1 h60636b9_2 conda-forge
freexl 2.0.0 h3ec172f_0 conda-forge
fribidi 1.0.10 hbcb3906_0 conda-forge
frozenlist 1.4.1 py312hb553811_1 conda-forge
fsspec 2024.9.0 pyhff2d567_0 conda-forge
gdal 3.9.1 py312h9b1be66_3 conda-forge
geobuf 1.1.1 pyh9f0ad1d_0 conda-forge
geographiclib 2.0 pypi_0 pypi
geopandas 1.0.1 pyhd8ed1ab_1 conda-forge
geopandas-base 1.0.1 pyha770c72_1 conda-forge
geopy 2.4.1 pypi_0 pypi
geos 3.12.1 h93d8f39_0 conda-forge
geotiff 1.7.3 h4bbec01_2 conda-forge
gettext 0.22.5 hdfe23c8_3 conda-forge
gettext-tools 0.22.5 hdfe23c8_3 conda-forge
gflags 2.2.2 hac325c4_1005 conda-forge
giflib 5.2.2 h10d778d_0 conda-forge
glog 0.7.1 h2790a97_0 conda-forge
gmp 6.3.0 hf036a51_2 conda-forge
gmpy2 2.1.5 py312h165121d_2 conda-forge
graphite2 1.3.13 h73e2aa4_1003 conda-forge
h11 0.14.0 pyhd8ed1ab_0 conda-forge
h2 4.1.0 pyhd8ed1ab_0 conda-forge
h5netcdf 1.3.0 pyhd8ed1ab_0 conda-forge
h5py 3.11.0 nompi_py312hfc94b03_102 conda-forge
harfbuzz 9.0.0 h098a298_1 conda-forge
hdf4 4.2.15 h8138101_7 conda-forge
hdf5 1.14.3 nompi_h687a608_105 conda-forge
hdf5plugin 5.0.0 py312h54c024f_0 conda-forge
holidays 0.57 pyhd8ed1ab_0 conda-forge
holoviews 1.19.1 pyhd8ed1ab_0 conda-forge
hpack 4.0.0 pyh9f0ad1d_0 conda-forge
html5lib 1.1 pyh9f0ad1d_0 conda-forge
httpcore 1.0.6 pyhd8ed1ab_0 conda-forge
httpx 0.27.2 pyhd8ed1ab_0 conda-forge
hvplot 0.11.0 pyhd8ed1ab_0 conda-forge
hyperframe 6.0.1 pyhd8ed1ab_0 conda-forge
icu 75.1 h120a0e1_0 conda-forge
idna 3.10 pyhd8ed1ab_0 conda-forge
importlib-metadata 8.5.0 pyha770c72_0 conda-forge
importlib-resources 6.4.5 pyhd8ed1ab_0 conda-forge
importlib_metadata 8.5.0 hd8ed1ab_0 conda-forge
importlib_resources 6.4.5 pyhd8ed1ab_0 conda-forge
ipykernel 6.29.5 pyh57ce528_0 conda-forge
ipython 8.28.0 pyh707e725_0 conda-forge
ipywidgets 8.1.5 pyhd8ed1ab_0 conda-forge
isoduration 20.11.0 pyhd8ed1ab_0 conda-forge
itsdangerous 2.2.0 pyhd8ed1ab_0 conda-forge
jasper 4.2.4 hb10263b_0 conda-forge
jdcal 1.4.1 py_0 conda-forge
jedi 0.19.1 pyhd8ed1ab_0 conda-forge
jinja2 3.1.4 pyhd8ed1ab_0 conda-forge
jiter 0.5.0 py312h669792a_1 conda-forge
jmespath 1.0.1 pyhd8ed1ab_0 conda-forge
joblib 1.4.2 pyhd8ed1ab_0 conda-forge
json-c 0.17 h6253ea5_1 conda-forge
json5 0.9.25 pyhd8ed1ab_0 conda-forge
jsonpickle 3.3.0 pyhd8ed1ab_0 conda-forge
jsonpointer 3.0.0 py312hb401068_1 conda-forge
jsonschema 4.23.0 pyhd8ed1ab_0 conda-forge
jsonschema-specifications 2023.12.1 pyhd8ed1ab_0 conda-forge
jsonschema-with-format-nongpl 4.23.0 hd8ed1ab_0 conda-forge
jupyter 1.1.1 pyhd8ed1ab_0 conda-forge
jupyter-lsp 2.2.5 pyhd8ed1ab_0 conda-forge
jupyter_client 8.6.3 pyhd8ed1ab_0 conda-forge
jupyter_console 6.6.3 pyhd8ed1ab_0 conda-forge
jupyter_core 5.7.2 pyh31011fe_1 conda-forge
jupyter_events 0.10.0 pyhd8ed1ab_0 conda-forge
jupyter_server 2.14.2 pyhd8ed1ab_0 conda-forge
jupyter_server_terminals 0.5.3 pyhd8ed1ab_0 conda-forge
jupyterlab 4.2.5 pyhd8ed1ab_0 conda-forge
jupyterlab_pygments 0.3.0 pyhd8ed1ab_1 conda-forge
jupyterlab_server 2.27.3 pyhd8ed1ab_0 conda-forge
jupyterlab_widgets 3.0.13 pyhd8ed1ab_0 conda-forge
kealib 1.5.3 he475af8_2 conda-forge
kiwisolver 1.4.7 py312hc5c4d5f_0 conda-forge
krb5 1.21.3 h37d8d59_0 conda-forge
lat_lon_parser 1.3.0 pyhd8ed1ab_0 conda-forge
lcms2 2.16 ha2f27b4_0 conda-forge
ld64_osx-64 609 h0fd476b_15 conda-forge
lerc 4.0.0 hb486fe8_0 conda-forge
libabseil 20240116.2 cxx17_hf036a51_1 conda-forge
libaec 1.1.3 h73e2aa4_0 conda-forge
libarchive 3.7.4 h20e244c_0 conda-forge
libarrow 17.0.0 ha60c65e_13_cpu conda-forge
libarrow-acero 17.0.0 hac325c4_13_cpu conda-forge
libarrow-dataset 17.0.0 hac325c4_13_cpu conda-forge
libarrow-flight 17.0.0 hea76c88_13_cpu conda-forge
libarrow-flight-sql 17.0.0 h824516f_13_cpu conda-forge
libarrow-gandiva 17.0.0 h8c10372_13_cpu conda-forge
libarrow-substrait 17.0.0 hba007a9_13_cpu conda-forge
libasprintf 0.22.5 hdfe23c8_3 conda-forge
libasprintf-devel 0.22.5 hdfe23c8_3 conda-forge
libblas 3.9.0 22_osx64_openblas conda-forge
libboost-headers 1.86.0 h694c41f_2 conda-forge
libbrotlicommon 1.1.0 h00291cd_2 conda-forge
libbrotlidec 1.1.0 h00291cd_2 conda-forge
libbrotlienc 1.1.0 h00291cd_2 conda-forge
libcblas 3.9.0 22_osx64_openblas conda-forge
libclang-cpp15 15.0.7 default_h7151d67_5 conda-forge
libcrc32c 1.1.2 he49afe7_0 conda-forge
libcurl 8.10.1 h58e7537_0 conda-forge
libcxx 19.1.1 hf95d169_0 conda-forge
libdeflate 1.20 h49d49c5_0 conda-forge
libedit 3.1.20191231 h0678c8f_2 conda-forge
libev 4.33 h10d778d_2 conda-forge
libevent 2.1.12 ha90c15b_1 conda-forge
libexpat 2.6.3 hac325c4_0 conda-forge
libffi 3.4.2 h0d85af4_5 conda-forge
libgdal 3.9.1 hb1a0af8_3 conda-forge
libgettextpo 0.22.5 hdfe23c8_3 conda-forge
libgettextpo-devel 0.22.5 hdfe23c8_3 conda-forge
libgfortran 5.0.0 13_2_0_h97931a8_3 conda-forge
libgfortran5 13.2.0 h2873a65_3 conda-forge
libglib 2.82.1 h63bbcf2_0 conda-forge
libgoogle-cloud 2.28.0 h721cda5_0 conda-forge
libgoogle-cloud-storage 2.28.0 h9e84e37_0 conda-forge
libgrpc 1.62.2 h384b2fc_0 conda-forge
libhwloc 2.11.1 default_h456cccd_1000 conda-forge
libiconv 1.17 hd75f5a5_2 conda-forge
libintl 0.22.5 hdfe23c8_3 conda-forge
libintl-devel 0.22.5 hdfe23c8_3 conda-forge
libjpeg-turbo 3.0.0 h0dc2134_1 conda-forge
libkml 1.3.0 h9ee1731_1021 conda-forge
liblapack 3.9.0 22_osx64_openblas conda-forge
libllvm14 14.0.6 hc8e404f_4 conda-forge
libllvm15 15.0.7 hbedff68_4 conda-forge
libllvm17 17.0.6 hbedff68_1 conda-forge
libllvm18 18.1.8 h9ce406d_2 conda-forge
libnetcdf 4.9.2 nompi_h7334405_114 conda-forge
libnghttp2 1.58.0 h64cf6d3_1 conda-forge
libntlm 1.4 h0d85af4_1002 conda-forge
libopenblas 0.3.27 openmp_h8869122_1 conda-forge
libparquet 17.0.0 hf1b0f52_13_cpu conda-forge
libpng 1.6.44 h4b8f8c9_0 conda-forge
libpq 16.4 h75a757a_2 conda-forge
libprotobuf 4.25.3 hd4aba4c_1 conda-forge
libre2-11 2023.09.01 h81f5012_2 conda-forge
librttopo 1.1.0 hf05f67e_15 conda-forge
libsodium 1.0.20 hfdf4475_0 conda-forge
libspatialindex 2.0.0 hf036a51_0 conda-forge
libspatialite 5.1.0 h5579707_7 conda-forge
libsqlite 3.46.1 h4b8f8c9_0 conda-forge
libssh2 1.11.0 hd019ec5_0 conda-forge
libthrift 0.20.0 h75589b3_1 conda-forge
libtiff 4.6.0 h129831d_3 conda-forge
libudunits2 2.2.28 h516ac8c_3 conda-forge
libutf8proc 2.8.0 hb7f2c08_0 conda-forge
libuuid 2.38.1 hb7f2c08_0 conda-forge
libwebp-base 1.4.0 h10d778d_0 conda-forge
libxcb 1.17.0 hf1f96e2_0 conda-forge
libxml2 2.12.7 heaf3512_4 conda-forge
libxslt 1.1.39 h03b04e6_0 conda-forge
libzip 1.11.1 h3116616_0 conda-forge
libzlib 1.3.1 hd23fc13_2 conda-forge
linkify-it-py 2.0.3 pyhd8ed1ab_0 conda-forge
llvm-openmp 19.1.0 h56322cc_0 conda-forge
llvm-tools 15.0.7 hbedff68_4 conda-forge
llvmlite 0.43.0 py312hcc8fd36_1 conda-forge
locket 1.0.0 pyhd8ed1ab_0 conda-forge
lunarcalendar 0.0.9 py_0 conda-forge
lxml 5.3.0 py312h4feaf87_1 conda-forge
lz4 4.3.3 py312h83408cd_1 conda-forge
lz4-c 1.9.4 hf0c8a7f_0 conda-forge
lzo 2.10 h10d778d_1001 conda-forge
magics 4.15.4 hbffab32_1 conda-forge
magics-python 1.5.8 pyhd8ed1ab_1 conda-forge
make 4.4.1 h00291cd_2 conda-forge
mapclassify 2.8.1 pyhd8ed1ab_0 conda-forge
markdown 3.6 pyhd8ed1ab_0 conda-forge
markdown-it-py 3.0.0 pyhd8ed1ab_0 conda-forge
markupsafe 2.1.5 py312hb553811_1 conda-forge
marshmallow 3.22.0 pyhd8ed1ab_0 conda-forge
matplotlib 3.8.4 py312hb401068_2 conda-forge
matplotlib-base 3.8.4 py312hb6d62fa_2 conda-forge
matplotlib-inline 0.1.7 pyhd8ed1ab_0 conda-forge
mdit-py-plugins 0.4.2 pyhd8ed1ab_0 conda-forge
mdurl 0.1.2 pyhd8ed1ab_0 conda-forge
measurement 3.2.0 py_1 conda-forge
metpy 1.6.3 pyhd8ed1ab_0 conda-forge
minizip 4.0.7 h62b0c8d_0 conda-forge
mistune 3.0.2 pyhd8ed1ab_0 conda-forge
mpc 1.3.1 h9d8efa1_1 conda-forge
mpfr 4.2.1 haed47dc_3 conda-forge
mpmath 1.3.0 pyhd8ed1ab_0 conda-forge
msgpack-python 1.1.0 py312hc5c4d5f_0 conda-forge
multidict 6.1.0 py312h9131086_0 conda-forge
multiprocess 0.70.16 py312hb553811_1 conda-forge
multiurl 0.3.1 pyhd8ed1ab_0 conda-forge
munkres 1.1.4 pyh9f0ad1d_0 conda-forge
nbclient 0.10.0 pyhd8ed1ab_0 conda-forge
nbconvert 7.16.4 hd8ed1ab_1 conda-forge
nbconvert-core 7.16.4 pyhd8ed1ab_1 conda-forge
nbconvert-pandoc 7.16.4 hd8ed1ab_1 conda-forge
nbformat 5.10.4 pyhd8ed1ab_0 conda-forge
ncurses 6.5 hf036a51_1 conda-forge
nest-asyncio 1.6.0 pyhd8ed1ab_0 conda-forge
netcdf4 1.7.1 nompi_py312h683d7b0_102 conda-forge
networkx 3.3 pyhd8ed1ab_1 conda-forge
notebook 7.2.2 pyhd8ed1ab_0 conda-forge
notebook-shim 0.2.4 pyhd8ed1ab_0 conda-forge
nspr 4.35 hea0b92c_0 conda-forge
nss 3.105 h3135457_0 conda-forge
numba 0.60.0 py312hc3b515d_0 conda-forge
numcodecs 0.13.0 py312h1171441_0 conda-forge
numpy 1.26.4 py312he3a82b2_0 conda-forge
ollama 0.1.17 cpu_he06a1bc_0 conda-forge
openai 1.51.0 pyhd8ed1ab_0 conda-forge
openjpeg 2.5.2 h7310d3a_0 conda-forge
openldap 2.6.8 hcd2896d_0 conda-forge
openssl 3.4.0 hd471939_0 conda-forge
orc 2.0.2 h22b2039_0 conda-forge
overrides 7.7.0 pyhd8ed1ab_0 conda-forge
owslib 0.31.0 pyhd8ed1ab_0 conda-forge
packaging 24.1 pyhd8ed1ab_0 conda-forge
pandas 2.2.3 py312h98e817e_1 conda-forge
pandoc 3.5 h694c41f_0 conda-forge
pandocfilters 1.5.0 pyhd8ed1ab_0 conda-forge
panel 1.5.2 pyhd8ed1ab_0 conda-forge
pango 1.54.0 h115fe74_2 conda-forge
param 2.1.1 pyhff2d567_0 conda-forge
parso 0.8.4 pyhd8ed1ab_0 conda-forge
partd 1.4.2 pyhd8ed1ab_0 conda-forge
patsy 0.5.6 pyhd8ed1ab_0 conda-forge
pcre2 10.44 h7634a1b_2 conda-forge
pexpect 4.9.0 pyhd8ed1ab_0 conda-forge
pickleshare 0.7.5 py_1003 conda-forge
pillow 10.4.0 py312h683ea77_1 conda-forge
pint 0.24.3 pyhd8ed1ab_0 conda-forge
pip 24.2 pyh8b19718_1 conda-forge
pixman 0.43.4 h73e2aa4_0 conda-forge
pkgutil-resolve-name 1.3.10 pyhd8ed1ab_1 conda-forge
platformdirs 4.3.6 pyhd8ed1ab_0 conda-forge
plotly 5.24.1 pyhd8ed1ab_0 conda-forge
polars 1.9.0 py312h088783b_0 conda-forge
pooch 1.8.2 pyhd8ed1ab_0 conda-forge
poppler 24.04.0 h0face88_0 conda-forge
poppler-data 0.4.12 hd8ed1ab_0 conda-forge
portalocker 2.10.1 py312hb401068_0 conda-forge
portion 2.5.0 pyhd8ed1ab_0 conda-forge
postgresql 16.4 h4b98a8f_2 conda-forge
proj 9.4.1 hf92c781_1 conda-forge
prometheus_client 0.21.0 pyhd8ed1ab_0 conda-forge
prompt-toolkit 3.0.48 pyha770c72_0 conda-forge
prompt_toolkit 3.0.48 hd8ed1ab_0 conda-forge
prophet 1.1.5 py312h3264805_1 conda-forge
protobuf 4.25.3 py312hd13efa9_1 conda-forge
psutil 6.0.0 py312hb553811_1 conda-forge
pthread-stubs 0.4 h00291cd_1002 conda-forge
ptyprocess 0.7.0 pyhd3deb0d_0 conda-forge
pure_eval 0.2.3 pyhd8ed1ab_0 conda-forge
pyarrow 17.0.0 py312h0be7463_1 conda-forge
pyarrow-core 17.0.0 py312h63b501a_1_cpu conda-forge
pyarrow-hotfix 0.6 pyhd8ed1ab_0 conda-forge
pycparser 2.22 pyhd8ed1ab_0 conda-forge
pydantic 2.9.2 pyhd8ed1ab_0 conda-forge
pydantic-core 2.23.4 py312h669792a_0 conda-forge
pydap 3.5 pyhd8ed1ab_0 conda-forge
pygments 2.18.0 pyhd8ed1ab_0 conda-forge
pykdtree 1.3.13 py312h3a11e2b_1 conda-forge
pymeeus 0.5.12 pyhd8ed1ab_0 conda-forge
pymongo 4.10.1 py312h5861a67_0 conda-forge
pyobjc-core 10.3.1 py312hab44e94_1 conda-forge
pyobjc-framework-cocoa 10.3.1 py312hab44e94_1 conda-forge
pyogrio 0.9.0 py312h43b3a95_0 conda-forge
pyorbital 1.8.3 pyhd8ed1ab_0 conda-forge
pyparsing 3.1.4 pyhd8ed1ab_0 conda-forge
pypdf 5.0.1 pyha770c72_0 conda-forge
pyproj 3.6.1 py312haf32e09_9 conda-forge
pyresample 1.30.0 py312h98e817e_0 conda-forge
pyshp 2.3.1 pyhd8ed1ab_0 conda-forge
pysocks 1.7.1 pyha2e5f31_6 conda-forge
pyspectral 0.13.5 pyhd8ed1ab_0 conda-forge
pystac 1.11.0 pyhd8ed1ab_0 conda-forge
python 3.12.7 h8f8b54e_0_cpython conda-forge
python-dateutil 2.9.0 pyhd8ed1ab_0 conda-forge
python-dotenv 1.0.1 pyhd8ed1ab_0 conda-forge
python-eccodes 2.37.0 py312h3a11e2b_0 conda-forge
python-fastjsonschema 2.20.0 pyhd8ed1ab_0 conda-forge
python-geotiepoints 1.7.4 py312h5dc8b90_0 conda-forge
python-json-logger 2.0.7 pyhd8ed1ab_0 conda-forge
python-tzdata 2024.2 pyhd8ed1ab_0 conda-forge
python_abi 3.12 5_cp312 conda-forge
pytz 2024.1 pyhd8ed1ab_0 conda-forge
pyviz_comms 3.0.3 pyhd8ed1ab_0 conda-forge
pyyaml 6.0.2 py312hb553811_1 conda-forge
pyzmq 26.2.0 py312h54d5c6a_2 conda-forge
qhull 2020.2 h3c5361c_5 conda-forge
qtconsole-base 5.6.0 pyha770c72_0 conda-forge
qtpy 2.4.1 pyhd8ed1ab_0 conda-forge
rapidfuzz 3.10.0 py312h5861a67_0 conda-forge
rasterio 1.3.10 py312h1c98354_4 conda-forge
re2 2023.09.01 hb168e87_2 conda-forge
readline 8.2 h9e318b2_1 conda-forge
referencing 0.35.1 pyhd8ed1ab_0 conda-forge
requests 2.32.3 pyhd8ed1ab_0 conda-forge
retrying 1.3.3 pyhd8ed1ab_3 conda-forge
rfc3339-validator 0.1.4 pyhd8ed1ab_0 conda-forge
rfc3986-validator 0.1.1 pyh9f0ad1d_0 conda-forge
rioxarray 0.17.0 pyhd8ed1ab_0 conda-forge
rpds-py 0.20.0 py312h669792a_1 conda-forge
rtree 1.3.0 py312hb560d21_2 conda-forge
s3transfer 0.10.2 pyhd8ed1ab_0 conda-forge
satpy 0.51.0 pyhd8ed1ab_0 conda-forge
scikit-learn 1.5.2 py312h9d777eb_1 conda-forge
scipy 1.14.1 py312he82a568_0 conda-forge
seaborn 0.13.2 hd8ed1ab_2 conda-forge
seaborn-base 0.13.2 pyhd8ed1ab_2 conda-forge
seawater 3.3.5 pyhd8ed1ab_0 conda-forge
semver 3.0.2 pyhd8ed1ab_0 conda-forge
send2trash 1.8.3 pyh31c8845_0 conda-forge
setuptools 75.1.0 pyhd8ed1ab_0 conda-forge
shapely 2.0.4 py312h3daf033_1 conda-forge
sigtool 0.1.3 h88f4db0_0 conda-forge
simplejson 3.19.3 py312hb553811_1 conda-forge
six 1.16.0 pyh6c4a22f_0 conda-forge
snappy 1.2.1 he1e6707_0 conda-forge
sniffio 1.3.1 pyhd8ed1ab_0 conda-forge
snuggs 1.4.7 pyhd8ed1ab_1 conda-forge
sortedcontainers 2.4.0 pyhd8ed1ab_0 conda-forge
soupsieve 2.5 pyhd8ed1ab_1 conda-forge
spdlog 1.14.1 h325aa07_1 conda-forge
sqlite 3.46.1 he26b093_0 conda-forge
stack_data 0.6.2 pyhd8ed1ab_0 conda-forge
stamina 24.3.0 pyhd8ed1ab_0 conda-forge
stanio 0.5.1 pyhd8ed1ab_0 conda-forge
statsmodels 0.14.4 py312h3a11e2b_0 conda-forge
sympy 1.13.3 pypyh2585a3b_103 conda-forge
tabulate 0.9.0 pyhd8ed1ab_1 conda-forge
tapi 1100.0.11 h9ce4665_0 conda-forge
tbb 2021.13.0 h37c8870_0 conda-forge
tbb-devel 2021.13.0 hf74753b_0 conda-forge
tblib 3.0.0 pyhd8ed1ab_0 conda-forge
tenacity 9.0.0 pyhd8ed1ab_0 conda-forge
terminado 0.18.1 pyh31c8845_0 conda-forge
threadpoolctl 3.5.0 pyhc1e730c_0 conda-forge
tiledb 2.24.2 h313d0e2_12 conda-forge
tinycss2 1.3.0 pyhd8ed1ab_0 conda-forge
tk 8.6.13 h1abcd95_1 conda-forge
tomli 2.0.2 pyhd8ed1ab_0 conda-forge
toolz 1.0.0 pyhd8ed1ab_0 conda-forge
tornado 6.4.1 py312hb553811_1 conda-forge
tqdm 4.66.5 pyhd8ed1ab_0 conda-forge
traitlets 5.14.3 pyhd8ed1ab_0 conda-forge
trollimage 1.25.0 py312h1171441_0 conda-forge
trollsift 0.5.1 pyhd8ed1ab_0 conda-forge
types-python-dateutil 2.9.0.20241003 pyhff2d567_0 conda-forge
typing-extensions 4.12.2 hd8ed1ab_0 conda-forge
typing_extensions 4.12.2 pyha770c72_0 conda-forge
typing_utils 0.1.0 pyhd8ed1ab_0 conda-forge
tzcode 2024b h00291cd_0 conda-forge
tzdata 2024b hc8b5060_0 conda-forge
tzfpy 0.15.6 py312h669792a_1 conda-forge
uc-micro-py 1.0.3 pyhd8ed1ab_0 conda-forge
udunits2 2.2.28 h516ac8c_3 conda-forge
unidecode 1.3.8 pyhd8ed1ab_0 conda-forge
uri-template 1.3.0 pyhd8ed1ab_0 conda-forge
uriparser 0.9.8 h6aefe2f_0 conda-forge
urllib3 2.2.3 pyhd8ed1ab_0 conda-forge
validators 0.22.0 pyhd8ed1ab_0 conda-forge
watchdog 5.0.3 py312hb553811_0 conda-forge
wcwidth 0.2.13 pyhd8ed1ab_0 conda-forge
webcolors 24.8.0 pyhd8ed1ab_0 conda-forge
webencodings 0.5.1 pyhd8ed1ab_2 conda-forge
webob 1.8.8 pyhd8ed1ab_0 conda-forge
websocket-client 1.8.0 pyhd8ed1ab_0 conda-forge
werkzeug 3.0.4 pyhd8ed1ab_0 conda-forge
wheel 0.44.0 pyhd8ed1ab_0 conda-forge
widgetsnbextension 4.0.13 pyhd8ed1ab_0 conda-forge
wradlib 2.1.1 pyhd8ed1ab_0 conda-forge
wrapt 1.16.0 py312hb553811_1 conda-forge
xarray 2024.9.0 pyhd8ed1ab_0 conda-forge
xarray-datatree 0.0.14 pyhd8ed1ab_0 conda-forge
xclim 0.52.2 pyhd8ed1ab_0 conda-forge
xerces-c 3.2.5 h197e74d_2 conda-forge
xlsx2csv 0.8.3 pyhd8ed1ab_0 conda-forge
xlsxwriter 3.2.0 pyhd8ed1ab_0 conda-forge
xmltodict 0.13.0 pyhd8ed1ab_0 conda-forge
xorg-libxau 1.0.11 h00291cd_1 conda-forge
xorg-libxdmcp 1.1.5 h00291cd_0 conda-forge
xradar 0.6.5 pyhd8ed1ab_0 conda-forge
xyzservices 2024.9.0 pyhd8ed1ab_0 conda-forge
xz 5.2.6 h775f41a_0 conda-forge
yamale 5.2.1 pyhca7485f_0 conda-forge
yaml 0.2.5 h0d85af4_2 conda-forge
yarl 1.13.1 py312hb553811_0 conda-forge
zarr 2.18.3 pyhd8ed1ab_0 conda-forge
zeromq 4.3.5 hb33e954_5 conda-forge
zict 3.0.0 pyhd8ed1ab_0 conda-forge
zipp 3.20.2 pyhd8ed1ab_0 conda-forge
zlib 1.3.1 hd23fc13_2 conda-forge
zstandard 0.23.0 py312h7122b0e_1 conda-forge
zstd 1.5.6 h915ae27_0 conda-forge
```
</details>
|
open
|
2024-11-21T09:19:33Z
|
2024-11-22T21:15:22Z
|
https://github.com/SciTools/cartopy/issues/2482
|
[
"Component: Geometry transforms"
] |
guidocioni
| 4
|
jonaswinkler/paperless-ng
|
django
| 1,548
|
[BUG] Consume stops after initial run
|
**Describe the bug**
I am importing documents from my scanner to the consume directory. I've noticed that files are not picked up until I restart the container (running in Docker). After a restart, all files present are consumed correctly. After that initial run, if I continue placing files in the consume directory, no more will be picked up for consumption.
**To Reproduce**
1. Stop Docker container
2. Place files in consume directory
3. Start Docker container
4. Watch them being consumed
5. Now place some additional files in the directory
6. Watch them being **not** consumed!
**Expected behavior**
All files placed in the directory should be consumed after a short delay.
**Webserver logs**
I've posted the logs just before the last consume started. After `12:10:23 [Q] INFO recycled worker Process-1:13` I placed additional files in the directory which are not being picked up.
```
[2022-01-14 12:10:06,952] [INFO] [paperless.consumer] Consuming IMG_20220114_0003.pdf
[2022-01-14 12:10:16,126] [INFO] [paperless.handlers] Assigning correspondent EnBW to 2021-10-19 IMG_20220114_0002
[2022-01-14 12:10:16,155] [INFO] [paperless.handlers] Detected 2 potential document types, so we've opted for Rechnung
[2022-01-14 12:10:16,161] [INFO] [paperless.handlers] Assigning document type Rechnung to 2021-10-19 EnBW IMG_20220114_0002
[2022-01-14 12:10:16,187] [INFO] [paperless.handlers] Tagging "2021-10-19 EnBW IMG_20220114_0002" with "Vertrag"
[2022-01-14 12:10:17,271] [INFO] [paperless.consumer] Document 2021-10-19 EnBW IMG_20220114_0002 consumption finished
12:10:17 [Q] INFO Process-1:12 stopped doing work
12:10:17 [Q] INFO Processed [IMG_20220114_0002.pdf]
12:10:17 [Q] INFO recycled worker Process-1:12
12:10:17 [Q] INFO Process-1:14 ready for work at 579
12:10:17 [Q] INFO Process-1:14 processing [eighteen-colorado-potato-india]
12:10:17 [Q] INFO Process-1:14 stopped doing work
12:10:17 [Q] INFO Processed [eighteen-colorado-potato-india]
12:10:18 [Q] INFO recycled worker Process-1:14
12:10:18 [Q] INFO Process-1:15 ready for work at 581
[2022-01-14 12:10:22,528] [INFO] [paperless.handlers] Assigning correspondent DHBW Karlsruhe to 2022-01-14 IMG_20220114_0003
[2022-01-14 12:10:22,939] [INFO] [paperless.consumer] Document 2022-01-14 DHBW Karlsruhe IMG_20220114_0003 consumption finished
12:10:22 [Q] INFO Process-1:13 stopped doing work
12:10:23 [Q] INFO Processed [IMG_20220114_0003.pdf]
12:10:23 [Q] INFO recycled worker Process-1:13
12:10:23 [Q] INFO Process-1:16 ready for work at 589
[2022-01-14 12:10:56 +0000] [40] [CRITICAL] WORKER TIMEOUT (pid:45)
[2022-01-14 12:10:56 +0000] [40] [WARNING] Worker with pid 45 was terminated due to signal 6
12:19:59 [Q] INFO Enqueued 1
12:19:59 [Q] INFO Process-1 created a task from schedule [Check all e-mail accounts]
12:19:59 [Q] INFO Process-1:15 processing [fifteen-four-oregon-ceiling]
12:19:59 [Q] INFO Process-1:15 stopped doing work
12:19:59 [Q] INFO Processed [fifteen-four-oregon-ceiling]
12:19:59 [Q] INFO recycled worker Process-1:15
12:19:59 [Q] INFO Process-1:17 ready for work at 597
12:30:01 [Q] INFO Enqueued 1
12:30:01 [Q] INFO Process-1 created a task from schedule [Check all e-mail accounts]
12:30:01 [Q] INFO Process-1:16 processing [batman-iowa-sweet-berlin]
12:30:01 [Q] INFO Process-1:16 stopped doing work
12:30:01 [Q] INFO Processed [batman-iowa-sweet-berlin]
12:30:02 [Q] INFO recycled worker Process-1:16
12:30:02 [Q] INFO Process-1:18 ready for work at 599
12:39:33 [Q] INFO Enqueued 1
12:39:33 [Q] INFO Process-1 created a task from schedule [Check all e-mail accounts]
12:39:33 [Q] INFO Process-1:17 processing [timing-emma-football-monkey]
12:39:34 [Q] INFO Process-1:17 stopped doing work
12:39:34 [Q] INFO Processed [timing-emma-football-monkey]
12:39:34 [Q] INFO recycled worker Process-1:17
12:39:34 [Q] INFO Process-1:19 ready for work at 601
12:49:36 [Q] INFO Enqueued 1
12:49:36 [Q] INFO Process-1 created a task from schedule [Check all e-mail accounts]
12:49:36 [Q] INFO Process-1:18 processing [leopard-kansas-pip-orange]
12:49:36 [Q] INFO Process-1:18 stopped doing work
12:49:36 [Q] INFO Processed [leopard-kansas-pip-orange]
12:49:37 [Q] INFO recycled worker Process-1:18
12:49:37 [Q] INFO Process-1:20 ready for work at 603
```
**Relevant information**
- Running in Docker (on Kubernetes)
- 1.4.5
|
closed
|
2022-01-14T13:02:36Z
|
2022-01-20T09:41:10Z
|
https://github.com/jonaswinkler/paperless-ng/issues/1548
|
[] |
PhilippCh
| 2
|
sinaptik-ai/pandas-ai
|
data-visualization
| 1,488
|
Local LLM pandasai.json
|
### System Info
"name": "pandasai-all",
"version": "1.0.0",
MacOS (15.1.1)
The code is run directly as Poetry run. and not in a docker.
### 🐛 Describe the bug
I'm trying to use a local LLM but it keeps defaulting to BamboLLM
Here how the pandasai.json at the root directory looks like
Regardless what "llm" variable i use it defaults to BambooLLM!
```
"llm": "LLM",
"llm_options": {
"model": "Llama-3.3-70B-Instruct",
"api_url": "http://localhost:9000/v1"
}
```
This is the supported list
```
__all__ = [
"LLM",
"BambooLLM",
"AzureOpenAI",
"OpenAI",
"GooglePalm",
"GoogleVertexAI",
"GoogleGemini",
"HuggingFaceTextGen",
"LangchainLLM",
"BedrockClaude",
"IBMwatsonx",
]
```
|
closed
|
2024-12-19T10:45:00Z
|
2025-01-20T10:09:51Z
|
https://github.com/sinaptik-ai/pandas-ai/issues/1488
|
[
"bug"
] |
ahadda5
| 7
|
slackapi/python-slack-sdk
|
asyncio
| 1,261
|
blocks/attachments as str for chat.* API calls should be clearly supported
|
The use case reported at https://github.com/slackapi/python-slack-sdk/pull/1259#issuecomment-1237007209 has been supported for a long time but it was **not by design**.
```python
client = WebClient(token="....")
client.chat_postMessage(text="fallback", blocks="{ JSON string here }")
```
The `blocks` and `attachments` arguments for chat.postMessage API etc. are supposed to be `Sequence[Block | Attachment | dict]` as of today. However, passing the whole blocks/attachments as a single str should be a relatively common use case. In future versions, the use case should be clearly covered in both type hints and its implementation.
### Category (place an `x` in each of the `[ ]`)
- [x] **slack_sdk.web.WebClient (sync/async)** (Web API client)
- [ ] **slack_sdk.webhook.WebhookClient (sync/async)** (Incoming Webhook, response_url sender)
- [ ] **slack_sdk.models** (UI component builders)
- [ ] **slack_sdk.oauth** (OAuth Flow Utilities)
- [ ] **slack_sdk.socket_mode** (Socket Mode client)
- [ ] **slack_sdk.audit_logs** (Audit Logs API client)
- [ ] **slack_sdk.scim** (SCIM API client)
- [ ] **slack_sdk.rtm** (RTM client)
- [ ] **slack_sdk.signature** (Request Signature Verifier)
### Requirements
Please read the [Contributing guidelines](https://github.com/slackapi/python-slack-sdk/blob/main/.github/contributing.md) and [Code of Conduct](https://slackhq.github.io/code-of-conduct) before creating this issue or pull request. By submitting, you are agreeing to those rules.
|
closed
|
2022-09-06T04:44:42Z
|
2022-09-06T06:41:40Z
|
https://github.com/slackapi/python-slack-sdk/issues/1261
|
[
"enhancement",
"web-client",
"Version: 3x"
] |
seratch
| 0
|
lk-geimfari/mimesis
|
pandas
| 1,583
|
Replace black with ruff.
|
Ruff would be a good fit for us.
|
open
|
2024-07-19T13:24:33Z
|
2024-07-19T13:25:11Z
|
https://github.com/lk-geimfari/mimesis/issues/1583
|
[] |
lk-geimfari
| 0
|
tensorflow/tensor2tensor
|
machine-learning
| 1,449
|
ImportError: No module named 'mesh_tensorflow.transformer'
|
### Description
Importing `t2t_trainer` is not working in `1.12.0`.
I have just updated my t2t version to `1.12.0` and noticed that I cannot import `t2t_trainer` anymor as I am getting
> `ImportError: No module named 'mesh_tensorflow.transformer'`
which was not the case in `1.11.0`.
...
### Reproduce
```python
from tensor2tensor.bin import t2t_trainer
if __name__ == '__main__':
t2t_trainer.main(None)
```
### Stack trace
```
Traceback (most recent call last):
File "/home/sfalk/tmp/pycharm_project_265/asr/punctuation/test.py", line 1, in <module>
from tensor2tensor.bin import t2t_trainer
File "/home/sfalk/miniconda3/envs/t2t/lib/python3.5/site-packages/tensor2tensor/bin/t2t_trainer.py", line 24, in <module>
from tensor2tensor import models # pylint: disable=unused-import
File "/home/sfalk/miniconda3/envs/t2t/lib/python3.5/site-packages/tensor2tensor/models/__init__.py", line 35, in <module>
from tensor2tensor.models import mtf_transformer2
File "/home/sfalk/miniconda3/envs/t2t/lib/python3.5/site-packages/tensor2tensor/models/mtf_transformer2.py", line 23, in <module>
from mesh_tensorflow.transformer import moe
ImportError: No module named 'mesh_tensorflow.transformer'
```
### Environment information
```
OS: Linux 4.15.0-45-generic #48~16.04.1-Ubuntu SMP Tue Jan 29 18:03:48 UTC 2019 x86_64 x86_64 x86_64 GNU/Linux
$ pip freeze | grep tensor
mesh-tensorflow==0.0.4
tensor2tensor==1.12.0
tensorboard==1.12.0
tensorflow-gpu==1.12.0
tensorflow-metadata==0.9.0
tensorflow-probability==0.5.0
$ python -V
Python 3.5.6 :: Anaconda, Inc.
```
|
closed
|
2019-02-13T08:41:16Z
|
2019-02-13T08:45:50Z
|
https://github.com/tensorflow/tensor2tensor/issues/1449
|
[] |
stefan-falk
| 1
|
microsoft/nni
|
machine-learning
| 5,431
|
Can not prune model TypeError: 'model' object is not iterable
|
**Describe the issue**:
**Environment**:
- NNI version: 2.6.1
- Training service (local|remote|pai|aml|etc):
- Client OS:
- Server OS (for remote mode only):
- Python version:
- PyTorch/TensorFlow version:
- Is conda/virtualenv/venv used?:
- Is running in Docker?:
**Configuration**:
- Experiment config (remember to remove secrets!):
- Search space:
**Log message**:
- nnimanager.log:
- dispatcher.log:
- nnictl stdout and stderr:
<!--
Where can you find the log files:
LOG: https://github.com/microsoft/nni/blob/master/docs/en_US/Tutorial/HowToDebug.md#experiment-root-director
STDOUT/STDERR: https://nni.readthedocs.io/en/stable/reference/nnictl.html#nnictl-log-stdout
-->
**How to reproduce it?**:
I have some simple model:
```
class get_model(nn.Module):
def __init__(self, args, num_channel=3, num_class=40, **kwargs):
super(get_model, self).__init__()
self.args = args
self.bn1 = nn.BatchNorm2d(64)
self.bn2 = nn.BatchNorm2d(64)
self.bn3 = nn.BatchNorm2d(128)
self.bn4 = nn.BatchNorm2d(256)
self.bn5 = nn.BatchNorm1d(args.emb_dims)
self.conv1 = nn.Sequential(nn.Conv2d(num_channel*2, 64, kernel_size=1, bias=False),
self.bn1,
nn.LeakyReLU(negative_slope=0.2))
self.conv2 = nn.Sequential(nn.Conv2d(64*2, 64, kernel_size=1, bias=False),
self.bn2,
nn.LeakyReLU(negative_slope=0.2))
self.conv3 = nn.Sequential(nn.Conv2d(64*2, 128, kernel_size=1, bias=False),
self.bn3,
nn.LeakyReLU(negative_slope=0.2))
self.conv4 = nn.Sequential(nn.Conv2d(128*2, 256, kernel_size=1, bias=False),
self.bn4,
nn.LeakyReLU(negative_slope=0.2))
self.conv5 = nn.Sequential(nn.Conv1d(512, args.emb_dims, kernel_size=1, bias=False),
self.bn5,
nn.LeakyReLU(negative_slope=0.2))
self.linear1 = nn.Linear(args.emb_dims*2, 512, bias=False)
self.bn6 = nn.BatchNorm1d(512)
self.dp1 = nn.Dropout(p=args.dropout)
self.linear2 = nn.Linear(512, 256)
self.bn7 = nn.BatchNorm1d(256)
self.dp2 = nn.Dropout(p=args.dropout)
self.linear3 = nn.Linear(256, num_class)
def forward(self, x):
batch_size = x.size()[0]
x = get_graph_feature(x, k=self.args.k)
x = self.conv1(x)
x1 = x.max(dim=-1, keepdim=False)[0]
x = get_graph_feature(x1, k=self.args.k)
x = self.conv2(x)
x2 = x.max(dim=-1, keepdim=False)[0]
x = get_graph_feature(x2, k=self.args.k)
x = self.conv3(x)
x3 = x.max(dim=-1, keepdim=False)[0]
x = get_graph_feature(x3, k=self.args.k)
x = self.conv4(x)
x4 = x.max(dim=-1, keepdim=False)[0]
x = torch.cat((x1, x2, x3, x4), dim=1)
x = self.conv5(x)
x1 = F.adaptive_max_pool1d(x, 1).view(batch_size, -1)
x2 = F.adaptive_avg_pool1d(x, 1).view(batch_size, -1)
x = torch.cat((x1, x2), 1)
x = F.leaky_relu(self.bn6(self.linear1(x)), negative_slope=0.2)
x = self.dp1(x)
x = F.leaky_relu(self.bn7(self.linear2(x)), negative_slope=0.2)
x = self.dp2(x)
x = self.linear3(x)
return x
```
And I want to prune with different pruners, for example:
```
from nni.algorithms.compression.pytorch.pruning import L2NormPruner
pruner = L2NormPruner(model, config_list)
masked_model, masks = pruner.compress()
```
But get the error:
```
Traceback (most recent call last):
File "nni_optim.py", line 73, in <module>
masked_model, masks = pruner.compress()
TypeError: 'model' object is not iterable
```
Although in doc is written that model should be torch..nn.Module.
That is the issue with my model?
|
closed
|
2023-03-10T07:29:46Z
|
2023-03-13T07:47:34Z
|
https://github.com/microsoft/nni/issues/5431
|
[] |
Kracozebr
| 5
|
TheAlgorithms/Python
|
python
| 12,217
|
Add a index priority queue to Data Structures
|
### Feature description
As there is no IPQ implementation in the standard python library, I wish to add one to TheAlgorithms.
|
closed
|
2024-10-21T04:59:16Z
|
2024-10-21T05:03:04Z
|
https://github.com/TheAlgorithms/Python/issues/12217
|
[
"enhancement"
] |
alessadroc
| 1
|
flairNLP/flair
|
nlp
| 2,859
|
module 'conllu' has no attribute TokenList
|

This did not happen till today . I have been using this basic code for 6 months and this bug is new and did not appear anytime before today.
Please resolve quickly
|
closed
|
2022-07-12T18:03:37Z
|
2022-11-14T15:03:52Z
|
https://github.com/flairNLP/flair/issues/2859
|
[
"bug"
] |
yash-rathore
| 10
|
CorentinJ/Real-Time-Voice-Cloning
|
deep-learning
| 992
|
Error in loading state_dict for SpeakerEncoder: size mismatch
|
Hi! I trained a synthesizer a month ago and I could synthesize my voice, too mechanical though, but now I got this error. How can I fix this?
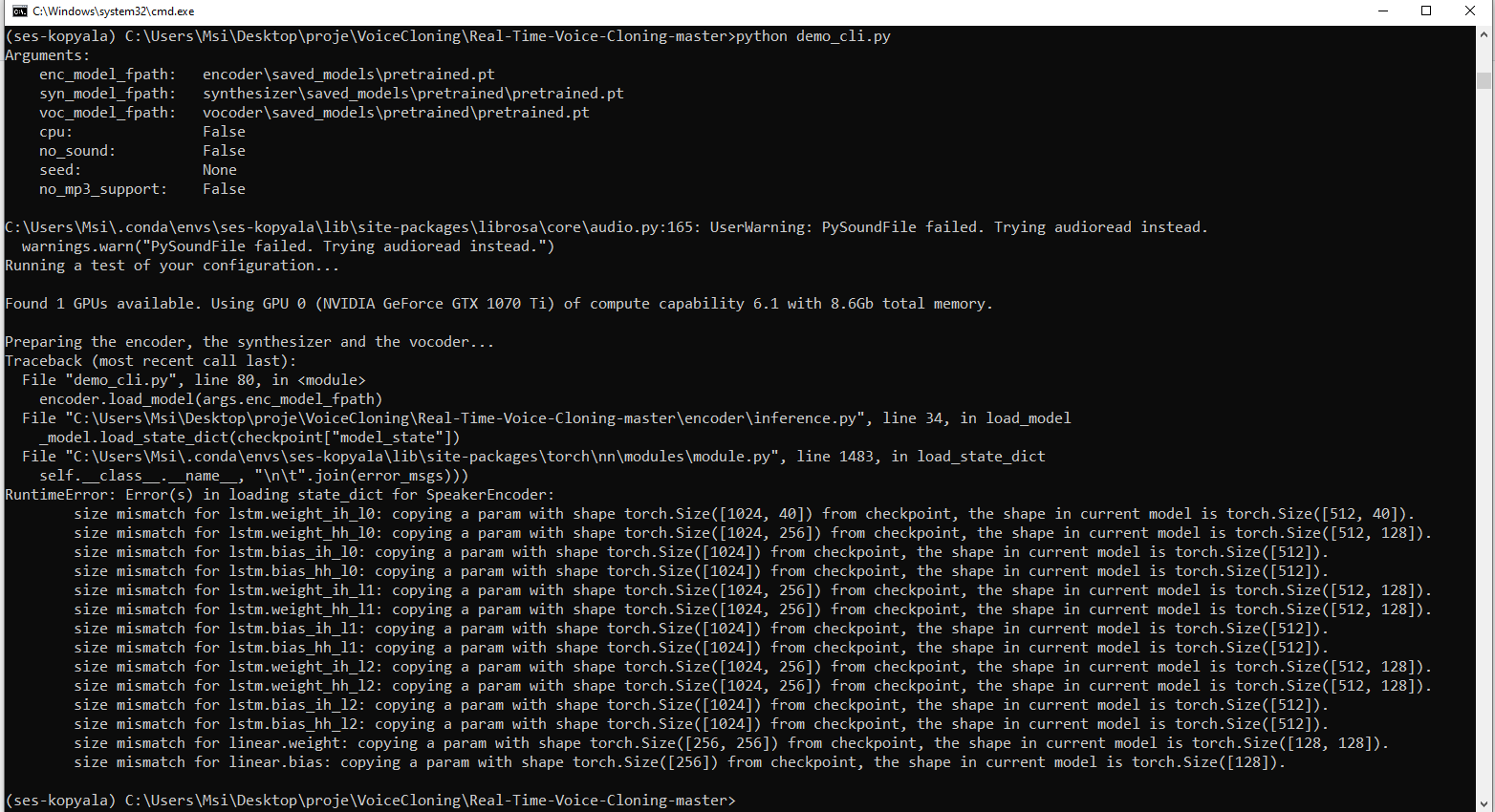
|
closed
|
2022-01-23T15:40:31Z
|
2022-01-28T19:41:32Z
|
https://github.com/CorentinJ/Real-Time-Voice-Cloning/issues/992
|
[] |
EkinUstundag
| 1
|
openapi-generators/openapi-python-client
|
fastapi
| 338
|
Add support for recursively defined schemas
|
**Describe the bug**
I tried to create a python client from an OpenApi-Spec with the command `openapi-python-client generate --path secret_server_openapi3.json`. Then I got multple warnings:
- invalid data in items of array settings
- Could not find reference in parsed models or enums
- Cannot parse response for status code 200, response will be ommitted from generated client
I searched the schemas, which are responsible for the errors and they all had in common, that they either reference another schema which is recursively defined or reference themself.
For example one of the problematic schemas:
```
{
"SecretDependencySetting": {
"type": "object",
"properties": {
"active": {
"type": "boolean",
"description": "Indicates the setting is active."
},
"childSettings": {
"type": "array",
"description": "The Child Settings that would be used for list of options.",
"items": {
"$ref": "#/components/schemas/SecretDependencySetting"
}
}
},
"description": "Secret Dependency Settings - Mostly used internally"
}
}
```
**To Reproduce**
Define a schema recursively and then try to create a client from it.
**Expected behavior**
The software can also parse a recursively defined schema.
**OpenAPI Spec File**
It's 66000 Lines, so I'm not sure if this will help you, or github will allow me to post it :smile:
Just ask if you need specific parts of the spec, aside from what I already provided above.
**Desktop (please complete the following information):**
- OS: Linux Manjaro
- Python Version: 3.9.1
- openapi-python-client version 0.7.3
|
closed
|
2021-02-17T13:27:58Z
|
2022-11-12T17:49:54Z
|
https://github.com/openapi-generators/openapi-python-client/issues/338
|
[
"✨ enhancement",
"🐲 here there be dragons"
] |
sp-schoen
| 13
|
WZMIAOMIAO/deep-learning-for-image-processing
|
deep-learning
| 730
|
FileNotFoundError: [Errno 2] No such file or directory: './save_weights/ssd300-14.pth'
|
有没有大哥可以分享一下训练好的ssd300-14.pth
|
open
|
2023-03-30T09:50:36Z
|
2023-03-30T09:50:36Z
|
https://github.com/WZMIAOMIAO/deep-learning-for-image-processing/issues/730
|
[] |
ANXIOUS-7
| 0
|
roboflow/supervision
|
tensorflow
| 1,044
|
[ByteTrack] add `minimum_consecutive_frames` to limit the number of falsely assigned tracker IDs
|
### Description
Expand the `ByteTrack` API by adding the `minimum_consecutive_frames` argument.
It will specify how many consecutive frames an object must be detected to be assigned a tracker ID. This will help prevent the creation of accidental tracker IDs in cases of false detection or double detection. Until detection reaches `minimum_consecutive_frames`, it should be assigned `-1`.
### API
```python
class ByteTrack:
def __init__(
self,
track_activation_threshold: float = 0.25,
lost_track_buffer: int = 30,
minimum_matching_threshold: float = 0.8,
frame_rate: int = 30,
minimum_consecutive_frames: int = 1
):
pass
```
### Additional
- Note: Please share a Google Colab with minimal code to test the new feature. We know it's additional work, but it will speed up the review process. The reviewer must test each change. Setting up a local environment to do this is time-consuming. Please ensure that Google Colab can be accessed without any issues (make it public). Thank you! 🙏🏻
|
closed
|
2024-03-25T15:29:06Z
|
2024-04-24T22:25:19Z
|
https://github.com/roboflow/supervision/issues/1044
|
[
"enhancement",
"api:tracker"
] |
SkalskiP
| 3
|
voila-dashboards/voila
|
jupyter
| 940
|
UI Tests: Update to the new Galata setup
|
<!--
Welcome! Thanks for thinking of a way to improve Voilà. If this solves a problem for you, then it probably solves that problem for lots of people! So the whole community will benefit from this request.
Before creating a new feature request please search the issues for relevant feature requests.
-->
### Problem
The new galata setup now lives in the core JupyterLab repo: https://github.com/jupyterlab/jupyterlab/tree/master/galata
We should update to it, as it comes with many improvements.
### Proposed Solution
Update the `ui-tests` setup here: https://github.com/voila-dashboards/voila/tree/master/ui-tests
Following the instructions here: https://github.com/jupyterlab/jupyterlab/tree/master/galata
<!-- Provide a clear and concise description of a way to accomplish what you want. For example:
* Add an option so that when [...] [...] will happen
-->
### Additional context
Galata was moved to the core lab repo in https://github.com/jupyterlab/jupyterlab/pull/10796
|
closed
|
2021-09-02T06:58:02Z
|
2021-09-03T16:02:02Z
|
https://github.com/voila-dashboards/voila/issues/940
|
[
"maintenance"
] |
jtpio
| 1
|
HumanSignal/labelImg
|
deep-learning
| 180
|
Image in portrait mode
|
I have some images which are took in portrait mode like this
https://imgur.com/baVUII8
But after I load the image, it becomes landscape mode
https://imgur.com/EQUGKIW
If it is in landscape mode, I cant label eyes and nose
both Win10 and Ubuntu16.04+python2+QT4 happened
How to fix this problem?
Thanks for ur helping
Best Wishes
|
closed
|
2017-10-23T06:25:10Z
|
2017-10-23T07:54:22Z
|
https://github.com/HumanSignal/labelImg/issues/180
|
[] |
nerv3890
| 1
|
mkhorasani/Streamlit-Authenticator
|
streamlit
| 10
|
Password modification
|
Great job !
Do you plan to add a "forgot password" option ?
|
closed
|
2022-05-16T13:32:30Z
|
2022-06-25T15:15:33Z
|
https://github.com/mkhorasani/Streamlit-Authenticator/issues/10
|
[] |
axel-prl-mrtg
| 2
|
neuml/txtai
|
nlp
| 41
|
Enhance API to fully support all txtai functionality
|
Currently, the API supports a subset of functionality in the embeddings module. Fully support embeddings and add methods qa extraction and labeling.
This will enable network-based implementations of txtai in other programming languages.
|
closed
|
2020-11-20T17:31:08Z
|
2021-05-13T15:04:12Z
|
https://github.com/neuml/txtai/issues/41
|
[] |
davidmezzetti
| 0
|
quantmind/pulsar
|
asyncio
| 195
|
Deprecate async function and replace with ensure_future
|
First step towards new syntax
|
closed
|
2016-01-30T20:38:38Z
|
2016-03-17T08:01:08Z
|
https://github.com/quantmind/pulsar/issues/195
|
[
"design decision",
"enhancement"
] |
lsbardel
| 1
|
learning-at-home/hivemind
|
asyncio
| 507
|
[BUG] Tests for compression fail on GPU servers with bitsandbytes installed
|
**Describe the bug**
While working on https://github.com/learning-at-home/hivemind/pull/490, I found that if I have bitsandbytes installed in a GPU-enabled environment, I get an error when running [test_adaptive_compression](https://github.com/learning-at-home/hivemind/blob/master/tests/test_compression.py#L152), which happens to be the only test that uses TrainingAverager under the hood.
I dug into it a bit, and the failure seems to be caused by `CUDA error: initialization error` from PyTorch, which AFAIK emerges when we're trying to initialize the CUDA context twice. More specifically, it appears when we are trying to initialize the optimizer states in TrainingAverager. My guess is that the context is created when importing bitsandbytes first and then when using something (anything?) from GPU-enabled PyTorch later. We are sunsetting the support for TrainingAverager anyway, but to me it's not obvious how to correctly migrate from this class in a given test.
**To Reproduce**
Install the environment in a GPU-enabled system, try running `CUDA_LAUNCH_BLOCKING=1 pytest -s --full-trace tests/test_compression.py`. Then uninstall bitsandbytes, comment out the parts in test_compression that rely on it (mostly `test_tensor_compression`), run the same command.
**Environment**
* Python 3.8.8
* [Commit 131f82c](https://github.com/learning-at-home/hivemind/commit/131f82c97ea67510d552bb7a68138ad27cbfa5d4)
* PyTorch 1.12.1, bitsandbytes 0.32.3
* NVIDIA RTX 2080 Ti GPU
|
open
|
2022-09-10T14:59:04Z
|
2022-09-10T14:59:04Z
|
https://github.com/learning-at-home/hivemind/issues/507
|
[
"bug",
"ci"
] |
mryab
| 0
|
dynaconf/dynaconf
|
fastapi
| 772
|
[bug] Setting auto_cast in instance options is ignored
|
**Affected version:** 3.1.9
**Describe the bug**
Setting **auto_cast** option inside `config.py` to **False** is ignored, even though the docs say otherwise.
**To Reproduce**
Add **auto_cast** to Dynaconf initialization.
```
from dynaconf import Dynaconf
settings = Dynaconf(
settings_files=["settings.toml", ".secrets.toml"],
auto_cast=False,
**more_options
)
```
Add the following to the `settings.toml`:
```
foo="@int 32"
```
Check value of **foo** in your `program.py`:
```
from config import settings
foo = settings.get('foo', False)
print(f"{foo=}", type(foo))
```
Executing `program.py` will return:
```
foo=32 <class 'int'>
```
when it should return:
```
foo='@int 32' <class 'str'>
```
Running `program.py` with **AUTO_CAST_FOR_DYNACONF=false** works as expected.
|
closed
|
2022-07-22T15:43:41Z
|
2022-09-22T12:49:36Z
|
https://github.com/dynaconf/dynaconf/issues/772
|
[
"bug"
] |
pvmm
| 1
|
Lightning-AI/pytorch-lightning
|
deep-learning
| 20,511
|
Cannot import OptimizerLRSchedulerConfig or OptimizerLRSchedulerConfigDict
|
### Bug description
Since I bumped up `lightning` to `2.5.0`, the `configure_optimizers` has been failing the type checker. I saw that `OptimizerLRSchedulerConfig` had been replaced with `OptimizerLRSchedulerConfigDict`, but I cannot import any of them.
### What version are you seeing the problem on?
v2.5
### How to reproduce the bug
```python
import torch
import pytorch_lightning as pl
from lightning.pytorch.utilities.types import OptimizerLRSchedulerConfigDict
from torch.optim.lr_scheduler import ReduceLROnPlateau
class Model(pl.LightningModule):
...
def configure_optimizers(self) -> OptimizerLRSchedulerConfigDict:
optimizer = torch.optim.Adam(self.parameters(), lr=1e-3)
scheduler = ReduceLROnPlateau(
optimizer, mode="min", factor=0.1, patience=20, min_lr=1e-6
)
return {
"optimizer": optimizer,
"lr_scheduler": {
"scheduler": scheduler,
"monitor": "val_loss",
"interval": "epoch",
"frequency": 1,
},
}
```
### Error messages and logs
```
In [2]: import lightning
In [3]: lightning.__version__
Out[3]: '2.5.0'
In [4]: from lightning.pytorch.utilities.types import OptimizerLRSchedulerConfigDict
---------------------------------------------------------------------------
ImportError Traceback (most recent call last)
Cell In[4], line 1
----> 1 from lightning.pytorch.utilities.types import OptimizerLRSchedulerConfigDict
ImportError: cannot import name 'OptimizerLRSchedulerConfigDict' from 'lightning.pytorch.utilities.types' (/home/test/.venv/lib/python3.11/site-packages/lightning/pytorch/utilities/types.py)
In [5]: from lightning.pytorch.utilities.types import OptimizerLRSchedulerConfig
---------------------------------------------------------------------------
ImportError Traceback (most recent call last)
Cell In[5], line 1
----> 1 from lightning.pytorch.utilities.types import OptimizerLRSchedulerConfig
ImportError: cannot import name 'OptimizerLRSchedulerConfig' from 'lightning.pytorch.utilities.types' (/home/test/.venv/lib/python3.11/site-packages/lightning/pytorch/utilities/types.py)
```
### Environment
<details>
<summary>Current environment</summary>
```
#- PyTorch Lightning Version (e.g., 2.5.0):
#- PyTorch Version (e.g., 2.5):
#- Python version (e.g., 3.12):
#- OS (e.g., Linux):
#- CUDA/cuDNN version:
#- GPU models and configuration:
#- How you installed Lightning(`conda`, `pip`, source):
```
</details>
### More info
_No response_
|
closed
|
2024-12-20T15:18:27Z
|
2024-12-21T01:42:58Z
|
https://github.com/Lightning-AI/pytorch-lightning/issues/20511
|
[
"bug",
"ver: 2.5.x"
] |
zordi-youngsun
| 4
|
iterative/dvc
|
machine-learning
| 10,452
|
Getting timeout in DVC pull
|
I have a large file arround 3GB
When I try to do dvc pull from inside a docker environment, I get this below error.
ERROR: unexpected error - The difference between the request time and the current time is too large.: An error occurred (RequestTimeTooSkewed) when calling the GetObject operation: The difference between the request time and the current time is too large.
Having any troubles? Hit us up at https://dvc.org/support, we are always happy to help!
Can you help regarding this ?
|
closed
|
2024-06-07T12:12:01Z
|
2025-01-12T15:10:47Z
|
https://github.com/iterative/dvc/issues/10452
|
[
"awaiting response",
"A: data-sync"
] |
bhaswa
| 6
|
minimaxir/textgenrnn
|
tensorflow
| 161
|
What version(s) of tensorflow are supported?
|
open
|
2019-12-14T03:32:11Z
|
2019-12-17T08:37:26Z
|
https://github.com/minimaxir/textgenrnn/issues/161
|
[] |
marcusturewicz
| 1
|
|
ymcui/Chinese-LLaMA-Alpaca
|
nlp
| 642
|
关于lora 精调 Alpaca-7B-Plus模型时为何不需要设置Lora参数的疑惑
|
### 提交前必须检查以下项目
- [X] 请确保使用的是仓库最新代码(git pull),一些问题已被解决和修复。
- [X] 由于相关依赖频繁更新,请确保按照[Wiki](https://github.com/ymcui/Chinese-LLaMA-Alpaca/wiki)中的相关步骤执行
- [X] 我已阅读[FAQ章节](https://github.com/ymcui/Chinese-LLaMA-Alpaca/wiki/常见问题)并且已在Issue中对问题进行了搜索,没有找到相似问题和解决方案
- [X] 第三方插件问题:例如[llama.cpp](https://github.com/ggerganov/llama.cpp)、[text-generation-webui](https://github.com/oobabooga/text-generation-webui)、[LlamaChat](https://github.com/alexrozanski/LlamaChat)等,同时建议到对应的项目中查找解决方案
- [X] 模型正确性检查:务必检查模型的[SHA256.md](https://github.com/ymcui/Chinese-LLaMA-Alpaca/blob/main/SHA256.md),模型不对的情况下无法保证效果和正常运行
### 问题类型
模型训练与精调
### 基础模型
Alpaca-Plus-7B
### 操作系统
Linux
### 详细描述问题
<img width="700" alt="image" src="https://github.com/ymcui/Chinese-LLaMA-Alpaca/assets/108610753/8ddc7cb5-9ae1-4ffb-a5e3-69f6ac9d7432">
一、我用您提供的指令训练了Chinese-Alpaca-7B-Plus的模型, 然而一部分较长的语句精调后能记住,而另一部分句子,则会发生遗忘。网上给出的建议是增加cut_off_length和增加lora权重,但是在您的指南里说精调Chinese-Alpaca-7B-Plus不需要设置Lora权重,请问这是为什么?
二、训练过程中我使用两张A5000(24GB),训练过程中计算loss函数的时候提示deepspeed OverFlow。但是测试训练结果的时候仍然比较令人满意,请问这里计算loss的时候的OverFlow是否可以直接忽略?
三、训练测试结果发现,lora算法,对于某些比较长的回答语句,会存在选择性记忆的情况,有些问题的回答可以做到与训练集一模一样,但有些问题的回答则是明显删减过的。请问这种情况下应当如何解决?我希望是回答得越精准越好,最好和我的训练集的答案一模一样。
非常感谢您的项目以及指南,对于我的工作有非常大的帮助!
### 依赖情况(代码类问题务必提供)
_No response_
### 运行日志或截图
_No response_
|
closed
|
2023-06-20T05:28:01Z
|
2023-06-27T23:56:01Z
|
https://github.com/ymcui/Chinese-LLaMA-Alpaca/issues/642
|
[
"stale"
] |
jjyu-ustc
| 2
|
marshmallow-code/marshmallow-sqlalchemy
|
sqlalchemy
| 232
|
Issue with primary keys that are also foreign keys
|
I have a case where marshmallow-sqlalchemy causes an SQLAlchemy FlushError at session commit time.
This is the error I get:
```
sqlalchemy.orm.exc.FlushError: New instance <Parent at 0x7f790f4832b0> with identity key (<class '__main__.Parent'>, (1,), None) conflicts with persistent instance <Parent at 0x7f790f4830f0>
```
My case is that of a Parent class/table with a primary key that also is a foreign key to a Child class/table.
The code that triggers the error is the following:
```py
parent = Session.query(Parent).one()
json_data = {"child": {"id": 1, "name": "new name"}}
with Session.no_autoflush:
instance = ParentSchema().load(json_data)
Session.add(instance.data)
Session.commit() # -> FlushError
```
`Session.query(Parent).one()` loads the parent object from the database and associates it with the session. `ParentSchema().load()` doesn't load the parent object from the database. Instead it creates a new object. And that new object has the same identity as the parent object that was loaded by `Session.query(Parent).one()`.
I tend to think that the problem is in [`ModelSchema.get_instance()`](https://github.com/marshmallow-code/marshmallow-sqlalchemy/blob/c46150667b98297a034dfe08582659129f9f9926/src/marshmallow_sqlalchemy/schema.py#L170-L182), which fails to load the instance from the database and return `None` instead.
This is the full test-case: https://gist.github.com/elemoine/dcd0475acb26cdbf827015c8fae744ba. The code that initially triggered this issue in my application is more complex than this. This test-case is the minimal code reproducing the issue that I've been able to come up with.
|
closed
|
2019-08-05T15:18:31Z
|
2025-01-12T05:37:10Z
|
https://github.com/marshmallow-code/marshmallow-sqlalchemy/issues/232
|
[] |
elemoine
| 3
|
dmlc/gluon-cv
|
computer-vision
| 865
|
Any channel pruning examples?
|
Hi there, I'm trying some channel pruning ideas in mxnet.
However it's hard to get some simple tutorials or examples I can start with.
So is there some basic examples showing how to do channel pruning (freezing)
during training?
An example of channel pruning is **Learning Efficient Convolutional Networks through Network Slimming** [arXiv](arxiv.org/abs/1708.06519) [PyTorch code](https://github.com/foolwood/pytorch-slimming).
And the example of channel freezing is **Soft Filter Pruning for Accelerating Deep Convolutional Neural Networks** [arXiv](arxiv.org/abs/1808.06866) [PyTorch code](https://github.com/he-y/soft-filter-pruning).
By channel pruning I mean eliminate some of the neurons (channels) during training.
By channel freezing I mean zero-mask the data (in forward) and the gradient (in backward) of part of the neurons during training.
I also raised a discussion here <https://discuss.gluon.ai/t/topic/13395>.
I just immigrated to MXNet from PyTorch so my main problem is that I'm not very familiar with
the API. For example I don't know how to access the data and gradient of model parameters.
____
Update-0:
I started to implement by myself. In the document of `Parameter` (<https://mxnet.incubator.apache.org/_modules/mxnet/gluon/parameter.html>), there only the
`set_data()` method. The `set_grad()` method is absent through which I can freeze some of the parameters by zeroing their gradients.
So how can I modify the parameter gradient manually?
|
closed
|
2019-07-14T09:50:16Z
|
2021-05-24T07:52:30Z
|
https://github.com/dmlc/gluon-cv/issues/865
|
[
"Stale"
] |
zeakey
| 1
|
geopandas/geopandas
|
pandas
| 3,512
|
BUG: to_arrow conversion should write crs as PROJJSON object and not string.
|
- [x] I have checked that this issue has not already been reported.
- [x] I have confirmed this bug exists on the latest version of geopandas.
- [ ] (optional) I have confirmed this bug exists on the main branch of geopandas.
---
#### Code Sample, a copy-pastable example
```python
import geopandas
import nanoarrow as na
df = geopandas.GeoDataFrame({"geometry": geopandas.GeoSeries.from_wkt(["POINT (0 1)"], crs="OGC:CRS84")})
na.Array(df.to_arrow()).schema.field(0).metadata
#> <nanoarrow._schema.SchemaMetadata>
#> - b'ARROW:extension:name': b'geoarrow.wkb'
#> - b'ARROW:extension:metadata': b'{"crs": "{\\"$schema\\":\\"https://proj.org/sch
```
#### Problem description
The `"crs"` field here is PROJJSON as a string within JSON (it should be a JSON object!). I suspect this is because somewhere there is a `to_json()` that maybe should be a `to_json_dict()`.
cc @jorisvandenbossche @kylebarron
#### Expected Output
#> - b'ARROW:extension:metadata': b'{"crs": {"$schema":"https://proj.org/sch
(i.e., value of CRS is a JSON object and not a string)
#### Output of ``geopandas.show_versions()``
<details>
SYSTEM INFO
-----------
python : 3.13.1 (main, Dec 3 2024, 17:59:52) [Clang 16.0.0 (clang-1600.0.26.4)]
executable : [/Users/dewey/gh/arrow/.venv/bin/python](https://file+.vscode-resource.vscode-cdn.net/Users/dewey/gh/arrow/.venv/bin/python)
machine : macOS-15.3-arm64-arm-64bit-Mach-O
GEOS, GDAL, PROJ INFO
---------------------
GEOS : 3.11.4
GEOS lib : None
GDAL : 3.9.1
GDAL data dir: [/Users/dewey/gh/arrow/.venv/lib/python3.13/site-packages/pyogrio/gdal_data/](https://file+.vscode-resource.vscode-cdn.net/Users/dewey/gh/arrow/.venv/lib/python3.13/site-packages/pyogrio/gdal_data/)
PROJ : 9.4.1
PROJ data dir: [/Users/dewey/gh/arrow/.venv/lib/python3.13/site-packages/pyproj/proj_dir/share/proj](https://file+.vscode-resource.vscode-cdn.net/Users/dewey/gh/arrow/.venv/lib/python3.13/site-packages/pyproj/proj_dir/share/proj)
PYTHON DEPENDENCIES
-------------------
geopandas : 1.0.1
numpy : 2.2.0
pandas : 2.2.3
pyproj : 3.7.0
shapely : 2.0.7
pyogrio : 0.10.0
geoalchemy2: None
geopy : None
matplotlib : None
mapclassify: None
fiona : None
psycopg : None
psycopg2 : None
pyarrow : 20.0.0.dev158+g8cb2868569.d20250213
</details>
|
closed
|
2025-02-13T05:30:49Z
|
2025-02-18T14:27:06Z
|
https://github.com/geopandas/geopandas/issues/3512
|
[
"bug"
] |
paleolimbot
| 1
|
davidsandberg/facenet
|
tensorflow
| 383
|
face authentication using custom dataset
|
Hi All,
I have trained set of images for which am able to create .pkl file. Now i need to authenticate with single image. Tried with existing code(classifier.py) but could not able to achieve the same.
Pls suggest the way forward
Thanks
vij
|
open
|
2017-07-18T15:43:44Z
|
2019-11-10T06:06:33Z
|
https://github.com/davidsandberg/facenet/issues/383
|
[] |
myinzack
| 2
|
piccolo-orm/piccolo
|
fastapi
| 593
|
Email column type
|
We don't have an email column type at the moment. It's because Postgres doesn't have an email column type.
However, I think it would be useful to designate a column as containing an email.
## Option 1
We can define a new column type, which basically just inherits from `Varchar`:
```python
class Email(Varchar):
pass
```
## Option 2
Or, let the user annotate a column as containing an email.
One option is using [Annotated](https://docs.python.org/3/library/typing.html#typing.Annotated), but the downside is it was only added in Python 3.9:
```python
from typing import Annotated
class MyTable(Table):
email: Annotated[Varchar, 'email'] = Varchar()
```
Or something like this instead:
```python
class MyTable(Table):
email = Varchar(validators=['email'])
```
## Benefits
The main reason it would be useful to have an email column is when using [create_pydantic_model](https://piccolo-orm.readthedocs.io/en/latest/piccolo/serialization/index.html#create-pydantic-model) to auto generate a Pydantic model, we can set the type to `EmailStr` (see [docs](https://pydantic-docs.helpmanual.io/usage/types/)).
## Input
Any input is welcome - once we decide on an approach, adding it will be pretty easy.
|
closed
|
2022-08-19T20:23:16Z
|
2022-08-20T21:42:11Z
|
https://github.com/piccolo-orm/piccolo/issues/593
|
[
"enhancement",
"good first issue",
"proposal - input needed"
] |
dantownsend
| 5
|
hbldh/bleak
|
asyncio
| 815
|
Distinguish Advertisement Data and Scan Response Data
|
* bleak version: 0.14.2
### Description
Could you clarify about a limitation of the library, is it possible to distinguish data, received by the first advertisement packet (PDU type: `ADV_IND`) and the subsequent scan response packet (PDU type: `SCAN_RSP`) from a specific device? Right now, launching the example https://github.com/hbldh/bleak/blob/develop/examples/discover.py, I see that it returns both advertisement data and scan response data joined together. I would like to see exactly which data were received through advertisement packet and which one through scan response packet. Is it possible to realize using Bleck library?
|
closed
|
2022-04-26T01:26:10Z
|
2022-04-26T07:42:13Z
|
https://github.com/hbldh/bleak/issues/815
|
[] |
RAlexeev
| 2
|
httpie/cli
|
python
| 518
|
h
|
o
|
closed
|
2016-09-12T07:11:16Z
|
2020-04-27T07:20:57Z
|
https://github.com/httpie/cli/issues/518
|
[] |
ghost
| 2
|
neuml/txtai
|
nlp
| 512
|
Add support for configurable text/object fields
|
Currently, the `text` and `object` fields are hardcoded throughout much of the code. This change will make the text and object fields configurable.
|
closed
|
2023-07-29T02:11:45Z
|
2023-07-29T02:17:45Z
|
https://github.com/neuml/txtai/issues/512
|
[] |
davidmezzetti
| 0
|
InstaPy/InstaPy
|
automation
| 6,147
|
Can't follow private accounts (even after enabling the option)
|
## Expected Behavior
I want to be able to follow private accounts, any help will be really appreciated!
## Current Behavior
I get this message: "is private account, by default skip"
Even after adding:
```
session.set_skip_users(skip_private=False,
skip_no_profile_pic=True,
no_profile_pic_percentage=100,
skip_business=True)
```
## Possible Solution (optional)
## InstaPy configuration
```
# imports
from instapy import InstaPy
from instapy import smart_run
# login credentials
insta_username = 'user'
insta_password = 'pass'
session = InstaPy(username=insta_username, password=insta_password)
with smart_run(session):
""" Activity flow """
# general settings
session.set_dont_include(['accounts'])
session.set_quota_supervisor(enabled=True,
sleep_after=["likes", "comments", "follows", "unfollows", "server_calls_h"],
sleepyhead=True,
stochastic_flow=True,
notify_me=True,
peak_likes_hourly=57,
peak_likes_daily=585,
peak_comments_hourly=21,
peak_comments_daily=182,
peak_follows_hourly=48,
peak_follows_daily=None,
peak_unfollows_hourly=35,
peak_unfollows_daily=402,
peak_server_calls_hourly=None,
peak_server_calls_daily=4700)
# follow
session.set_do_follow(enabled=True, percentage=35, times=1)
session.unfollow_users(amount=60, InstapyFollowed=(True, "nonfollowers"), style="FIFO", unfollow_after=90*60*60, sleep_delay=501)
# like
session.set_do_like(True, percentage=70)
session.set_delimit_liking(enabled=True, max_likes=100, min_likes=0)
session.set_dont_unfollow_active_users(enabled=True, posts=5)
session.set_relationship_bounds(enabled=True,
potency_ratio=None,
delimit_by_numbers=True,
max_followers=3000,
max_following=4490,
min_followers=100,
min_following=56,
min_posts=10,
max_posts=1000)
session.follow_likers(['accounts'], photos_grab_amount=1, follow_likers_per_photo=5, randomize=True, sleep_delay=200, interact=True)
session.set_skip_users(skip_private=False,
skip_no_profile_pic=True,
no_profile_pic_percentage=100,
skip_business=True)
# Commenting
session.set_do_comment(enabled=False)
```
|
closed
|
2021-04-12T17:50:17Z
|
2021-07-21T05:18:53Z
|
https://github.com/InstaPy/InstaPy/issues/6147
|
[
"wontfix"
] |
hecontreraso
| 1
|
gradio-app/gradio
|
deep-learning
| 10,252
|
Browser get Out of Memory when using Plotly for plotting.
|
### Describe the bug
I used Gradio to create a page for monitoring an image that needs to be refreshed continuously. When I used Plotly for plotting and set the refresh rate to 10 Hz, the browser showed an "**Out of Memory**" error after running for less than 10 minutes.
I found that the issue is caused by the `Plot.svelte` file generating new CSS, which is then continuously duplicated by `PlotlyPlot.svelte`.
This problem can be resolved by making the following change in the `Plot.svelte` file:
Replace:
```javascript
key += 1;
let type = value?.type;
```
With:
```javascript
let type = value?.type;
if (type !== "plotly") {
key += 1;
}
```
In other words, if the plot type is `plotly`, no new CSS will be generated.
Finally, I’m new to both Svelte and TypeScript, so some of my descriptions might not be entirely accurate, but this method does resolve the issue.
### Have you searched existing issues? 🔎
- [X] I have searched and found no existing issues
### Reproduction
```python
import gradio as gr
import numpy as np
from datetime import datetime
import plotly.express as px
def get_image(shape):
# data = caget(pv)
x = np.arange(0, shape[1])
y = np.arange(0, shape[0])
X, Y = np.meshgrid(x, y)
xc = np.random.randint(0, shape[1])
yc = np.random.randint(0, shape[0])
data = np.exp(-((X - xc) ** 2 + (Y - yc) ** 2) / (2 * 100**2)) * 1000
data = data.reshape(shape)
return data
fig: None = px.imshow(
get_image((1200, 1920)),
color_continuous_scale="jet",
)
fig["layout"]["uirevision"] = 'constant'
# fig["config"]["plotGlPixelRatio"] = 1
# fig.update_traces(hovertemplate="x: %{x} <br> y: %{y} <br> z: %{z} <br> color: %{color}")
# fig.update_layout(coloraxis_showscale=False)
fig["layout"]['hovermode']=False
fig["layout"]["annotations"]=None
def make_plot(width, height):
shape = (int(height), int(width))
img = get_image(shape)
## image plot
fig["data"][0].update(z=img)
return fig
with gr.Blocks(delete_cache=(120, 180)) as demo:
timer = gr.Timer(0.5, active=False)
with gr.Row():
with gr.Column(scale=1) as Column1:
with gr.Row():
shape_x = gr.Number(value=480, label="Width")
shape_y = gr.Number(value=320, label="Height")
with gr.Row():
start_btn = gr.Button(value="Start")
stop_btn = gr.Button(value="Stop")
with gr.Column(scale=2):
plot = gr.Plot(value=fig, label="Plot")
timer.tick(make_plot, inputs=[shape_x, shape_y], outputs=[plot])
stop_btn.click(
lambda: gr.Timer(active=False),
inputs=None,
outputs=[timer],
)
start_btn.click(
lambda: gr.Timer(0.1, active=True),
inputs=None,
outputs=[timer],
)
if __name__ == "__main__":
demo.queue(max_size=10, default_concurrency_limit=10)
demo.launch(server_name="0.0.0.0", server_port=8080, share=False, max_threads=30)
```
### Screenshot

### Logs
_No response_
### System Info
```shell
$ gradio environment
Gradio Environment Information:
------------------------------
Operating System: Linux
gradio version: 5.9.1
gradio_client version: 1.5.2
------------------------------------------------
gradio dependencies in your environment:
aiofiles: 23.2.1
anyio: 4.6.2.post1
audioop-lts is not installed.
fastapi: 0.115.4
ffmpy: 0.4.0
gradio-client==1.5.2 is not installed.
httpx: 0.27.2
huggingface-hub: 0.26.2
jinja2: 3.1.4
markupsafe: 2.1.5
numpy: 2.0.2
orjson: 3.10.11
packaging: 24.2
pandas: 2.2.3
pillow: 11.0.0
pydantic: 2.9.2
pydub: 0.25.1
python-multipart: 0.0.20
pyyaml: 6.0.2
ruff: 0.7.2
safehttpx: 0.1.6
semantic-version: 2.10.0
starlette: 0.41.2
tomlkit: 0.12.0
typer: 0.12.5
typing-extensions: 4.11.0
urllib3: 2.2.3
uvicorn: 0.32.0
authlib; extra == 'oauth' is not installed.
itsdangerous; extra == 'oauth' is not installed.
gradio_client dependencies in your environment:
fsspec: 2024.10.0
httpx: 0.27.2
huggingface-hub: 0.26.2
packaging: 24.2
typing-extensions: 4.11.0
websockets: 12.0
```
### Severity
Blocking usage of gradio
|
closed
|
2024-12-25T05:16:18Z
|
2025-02-08T00:56:23Z
|
https://github.com/gradio-app/gradio/issues/10252
|
[
"bug"
] |
Reflux00
| 0
|
KaiyangZhou/deep-person-reid
|
computer-vision
| 351
|
significant GPU memory cost on PRID
|
Here is my configuration.
```python
model:
name: 'resnet50'
pretrained: True
data:
type: 'video'
sources: ['prid2011']
targets: ['prid2011']
height: 224
width: 112
combineall: False
transforms: ['random_flip']
save_dir: 'log/osnet_x1_0_prid2011_softmax_cosinelr'
loss:
name: 'softmax'
softmax:
label_smooth: True
train:
optim: 'amsgrad'
lr: 0.0015
max_epoch: 250
batch_size: 8
fixbase_epoch: 0
open_layers: ['classifier']
lr_scheduler: 'cosine'
test:
batch_size: 300
dist_metric: 'euclidean'
normalize_feature: False
evaluate: False
eval_freq: -1
rerank: False
```
Batch size is 8 and the GPU memory usage is 7249M/12G on my computer with one GTX 1080 Ti (12GB).
seq_len have been shorten to 4. when the batch size is set to 16, the problem, out of memory, happens.
Another code, [Video-Person-Re-ID-Fantastic-Techniques-and-Where-to-Find-Them](https://github.com/ppriyank/Video-Person-Re-ID-Fantastic-Techniques-and-Where-to-Find-Them), is used to do the same task, i.e., video reid.
For the same configuration, e.g., ResNet50, image size of 224x112, batch size of 8, seq_len of 4, the memory usage is 7849M/12G. When the batch size is set to 32, the memory usage becomes 9869M/12G.
The pytorch versions of them are 1.1.0 and 1.3.0 respectively. However, this may be not the reason for the dramatically increase of memory usage.
Anyone knows why this situation happens? I'm very appreciate for any responses!
|
open
|
2020-07-03T11:51:54Z
|
2020-07-04T14:37:56Z
|
https://github.com/KaiyangZhou/deep-person-reid/issues/351
|
[] |
deropty
| 1
|
RafaelMiquelino/dash-flask-login
|
plotly
| 5
|
current_user.is_authenticated returns false in deployment
|
Hello,
Thanks for making this repository.
I have been using it with success on a localhost, but as soon as I deploy it, on a hosted server, the user authentication stops behaving. As the user logs in, it registers that the user is authenticated, but within less than a 1s the bool current_user.is_authenticated is set to false.
I have tried everything, and this problem is consistent for my code, that includes the code from this repository, and if one puts this repository on a server and runs it.
Thanks and all the best,
Max H
|
closed
|
2020-06-04T07:39:04Z
|
2020-06-09T09:23:49Z
|
https://github.com/RafaelMiquelino/dash-flask-login/issues/5
|
[] |
max454545
| 12
|
3b1b/manim
|
python
| 1,498
|
AnimationGroup height visual issue
|
I am trying to create a cool effect where the text is getting written and zoom in at the same time.
I tried to use the `AnimationGroup` object for that but this is how it renders :
https://user-images.githubusercontent.com/2827383/115975871-e9a4b700-a568-11eb-8194-fed4927fcfd2.mp4
First, the letters appear really distant from each other, small and also the height animation is being cut off, the letters come back to the original size.
The code is very simple :
```python
class KoreanWord(Scene):
def construct(self):
t = Text('실망')
group = []
group.append(Write(t))
group.append(t.animate.set_height(2))
self.play(AnimationGroup(*group, run_time=10))
```
Am I doing something wrong, or is it an issue?
Thanks in advance for your help.
|
open
|
2021-04-24T23:55:57Z
|
2021-06-17T00:43:57Z
|
https://github.com/3b1b/manim/issues/1498
|
[] |
vdegenne
| 1
|
graphql-python/graphene
|
graphql
| 1,361
|
Interfaces are ignored when define Mutation.
|
Hello.
First of all, thanks for all graphene developers and contributors.
* **What is the current behavior?**
Interface is ignored when define mutation. This is my code.
```python
class CreatePlan(Mutation):
class Meta:
interfaces = (PlanInterface, )
class Arguments:
name = String(required=True)
comment = String(default_value='')
goal = String(required=True)
start_date = DateTime(required=True)
end_date = DateTime(required=True)
def mutate(parent, info, **kwargs):
~~~
# this raises AssertionError:
# CreatePlan fields must be a mapping (dict / OrderedDict)
# with field names as keys or a function which returns such a mapping.
```
* **What is the expected behavior?**
I think Mutation should includes all interfaces and fields together.
* **Please tell us about your environment:**
Here is my environment.
- Version: 2.1.9
- Platform: Mac OS
* **Other information** (e.g. detailed explanation, stacktraces, related issues, suggestions how to fix, links for us to have context, eg. stackoverflow)
I think removing just a line can resolve this problem.
**branch v2**, graphene/types/mutation.py line 69
```python
@classmethod
def __init_subclass_with_meta__(
cls,
interfaces=(),
resolver=None,
output=None,
arguments=None,
_meta=None,
**options
):
if not _meta:
_meta = MutationOptions(cls)
output = output or getattr(cls, "Output", None)
fields = {}
for interface in interfaces:
assert issubclass(interface, Interface), (
'All interfaces of {} must be a subclass of Interface. Received "{}".'
).format(cls.__name__, interface)
fields.update(interface._meta.fields)
if not output:
# If output is defined, we don't need to get the fields
fields = OrderedDict(). # This ignores interfaces. Remove this.
for base in reversed(cls.__mro__):
fields.update(yank_fields_from_attrs(base.__dict__, _as=Field))
output = cls
```
Also, I found same problem on master branch.
```python
@classmethod
def __init_subclass_with_meta__(
cls,
interfaces=(),
resolver=None,
output=None,
arguments=None,
_meta=None,
**options,
):
if not _meta:
_meta = MutationOptions(cls)
output = output or getattr(cls, "Output", None)
fields = {}
for interface in interfaces:
assert issubclass(
interface, Interface
), f'All interfaces of {cls.__name__} must be a subclass of Interface. Received "{interface}".'
fields.update(interface._meta.fields)
if not output:
# If output is defined, we don't need to get the fields
fields = {}. # This ignores interfaces. Remove this.
for base in reversed(cls.__mro__):
fields.update(yank_fields_from_attrs(base.__dict__, _as=Field))
output = cls
```
|
open
|
2021-09-03T07:13:50Z
|
2021-09-03T07:13:50Z
|
https://github.com/graphql-python/graphene/issues/1361
|
[
"🐛 bug"
] |
maintain1210
| 0
|
aio-libs/aiomysql
|
sqlalchemy
| 203
|
Mistake :(
|
closed
|
2017-08-07T17:33:19Z
|
2017-08-07T17:39:54Z
|
https://github.com/aio-libs/aiomysql/issues/203
|
[] |
paccorsi
| 0
|
|
yihong0618/running_page
|
data-visualization
| 710
|
小米运动健康App导出的TCX文件结构与TCXReader解析格式不兼容,导致无法获取心率信息
|
Edit: tcx同步到strava
小米运动健康导出的tcx结构:
```
<TrainingCenterDatabase creator="Mi Fitness" version="1.0" xsi:schemaLocation="http://www.garmin.com/xmlschemas/TrainingCenterDatabase/v2 http://www.garmin.com/xmlschemas/TrainingCenterDatabasev2.xsd">
<script/>
<Activities>
<Activity Sport="">
<Id>2024-09-08T19:08:07.000Z</Id>
<Lap>
<TotalTimeSeconds>2425</TotalTimeSeconds>
<DistanceMeters>5178</DistanceMeters>
<Calories>198</Calories>
<HeartRateBpm>135</HeartRateBpm>
<Steps>6860</Steps>
<Track>
<Trackpoint>
<Time>2024-09-08T19:08:06.000Z</Time>
<Position>
<LatitudeDegrees>30.681241989135742</LatitudeDegrees>
<LongitudeDegrees>103.96613311767578</LongitudeDegrees>
</Position>
</Trackpoint>
...
</Track>
</Lap>
</Activity>
</Activities>
</TrainingCenterDatabase>
```
在这里,\<HeartRateBpm>135\</HeartRateBpm>对应app里的平均心率。
https://github.com/alenrajsp/tcxreader (代码里的import TCXReadert是这个吧)的示例tcx文件结构:
```
<Trackpoint>
<Time>2020-12-26T15:50:21.000Z</Time>
<Position>
<LatitudeDegrees>46.49732105433941</LatitudeDegrees>
<LongitudeDegrees>15.496849408373237</LongitudeDegrees>
</Position>
<AltitudeMeters>2277.39990234375</AltitudeMeters>
<DistanceMeters>5001.52978515625</DistanceMeters>
<HeartRateBpm>
<Value>148</Value>
</HeartRateBpm>
<Extensions>
<ns3:TPX>
<ns3:Speed>3.3589999675750732</ns3:Speed>
<ns3:RunCadence>61</ns3:RunCadence>
</ns3:TPX>
</Extensions>
</Trackpoint>
```
在这里,\<HeartRateBpm>包含一个\<Value>标签来存储心率数据而且在\<Trackpoint>里面。
不会改代码。。尝试在小米导出的tcx文件第一个\<Trackpoint>里加上对应\<HeartRateBpm>字段确实能够读到心率数据。。。
```
<Track>
<Trackpoint>
<Time>2024-09-08T19:08:06.000Z</Time>
<Position>
<LatitudeDegrees>30.681241989135742</LatitudeDegrees>
<LongitudeDegrees>103.96613311767578</LongitudeDegrees>
</Position>
<HeartRateBpm>
<Value>135</Value>
</HeartRateBpm>
</Trackpoint>
```

不知道这个问题能解决不。。
THx
|
closed
|
2024-09-08T15:02:34Z
|
2024-11-10T12:06:00Z
|
https://github.com/yihong0618/running_page/issues/710
|
[] |
RiverOnVenus
| 11
|
pydata/pandas-datareader
|
pandas
| 197
|
Docs bug
|
The [usage examples](http://pandas-datareader.readthedocs.org/en/latest/remote_data.html#yahoo-finance-options) for Yahoo Finance options contain a couple errors.
|
closed
|
2016-04-24T09:22:19Z
|
2016-04-24T09:55:49Z
|
https://github.com/pydata/pandas-datareader/issues/197
|
[] |
andportnoy
| 3
|
Textualize/rich
|
python
| 2,572
|
[BUG] Hyperlinks in logger messages don't work on VS Code
|
You may find a solution to your problem in the [docs](https://rich.readthedocs.io/en/latest/introduction.html) or [issues](https://github.com/textualize/rich/issues).
**Describe the bug**
The hyperlinks in the logger messages don't work in VS Code.
**Platform**
<details>
<summary>Click to expand</summary>
What platform (Win/Linux/Mac) are you running on? What terminal software are you using?
Linux
I may ask you to copy and paste the output of the following commands. It may save some time if you do it now.
If you're using Rich in a terminal:
```
python -m rich.diagnose
pip freeze | grep rich
```
If you're using Rich in a Jupyter Notebook, run the following snippet in a cell
and paste the output in your bug report.
```python
from rich.diagnose import report
report()
```


</details>
|
closed
|
2022-10-11T14:10:41Z
|
2024-08-26T14:29:55Z
|
https://github.com/Textualize/rich/issues/2572
|
[
"Needs triage"
] |
ma-sadeghi
| 3
|
dask/dask
|
numpy
| 11,574
|
Masked Array reductions and numpy 2+
|
<!-- Please include a self-contained copy-pastable example that generates the issue if possible.
Please be concise with code posted. See guidelines below on how to provide a good bug report:
- Craft Minimal Bug Reports http://matthewrocklin.com/blog/work/2018/02/28/minimal-bug-reports
- Minimal Complete Verifiable Examples https://stackoverflow.com/help/mcve
Bug reports that follow these guidelines are easier to diagnose, and so are often handled much more quickly.
-->
**Describe the issue**:
Some operations on masked arrays fail when using `numpy>2`. This is due to bad defaults present in numpy code for the `fill_value` of masked array, see https://github.com/numpy/numpy/issues/25677 and https://github.com/numpy/numpy/issues/27588
**Minimal Complete Verifiable Example**:
```python
import dask.array as da
xx = da.arange(0, 10, dtype='uint8')
xx = da.ma.masked_where(xx == 1, xx)
da.max(xx).compute()
```
Produces the following error:
<details>
<summary>Traceback</summary>
```python
---------------------------------------------------------------------------
TypeError Traceback (most recent call last)
Cell In[6], line 5
3 xx = da.ma.masked_where(xx == 1, xx)
----> 4 da.max(xx).compute()
File ~/miniconda3/envs/np21/lib/python3.13/site-packages/dask/base.py:372, in DaskMethodsMixin.compute(self, **kwargs)
348 def compute(self, **kwargs):
349 """Compute this dask collection
350
351 This turns a lazy Dask collection into its in-memory equivalent.
(...)
370 dask.compute
371 """
--> 372 (result,) = compute(self, traverse=False, **kwargs)
373 return result
File ~/miniconda3/envs/np21/lib/python3.13/site-packages/dask/base.py:660, in compute(traverse, optimize_graph, scheduler, get, *args, **kwargs)
657 postcomputes.append(x.__dask_postcompute__())
659 with shorten_traceback():
--> 660 results = schedule(dsk, keys, **kwargs)
662 return repack([f(r, *a) for r, (f, a) in zip(results, postcomputes)])
File ~/miniconda3/envs/np21/lib/python3.13/site-packages/dask/array/reductions.py:477, in chunk_max(x, axis, keepdims)
475 return array_safe([], x, ndmin=x.ndim, dtype=x.dtype)
476 else:
--> 477 return np.max(x, axis=axis, keepdims=keepdims)
File ~/miniconda3/envs/np21/lib/python3.13/site-packages/numpy/_core/fromnumeric.py:3199, in max(a, axis, out, keepdims, initial, where)
3080 @array_function_dispatch(_max_dispatcher)
3081 @set_module('numpy')
3082 def max(a, axis=None, out=None, keepdims=np._NoValue, initial=np._NoValue,
3083 where=np._NoValue):
3084 """
3085 Return the maximum of an array or maximum along an axis.
3086
(...)
3197 5
3198 """
-> 3199 return _wrapreduction(a, np.maximum, 'max', axis, None, out,
3200 keepdims=keepdims, initial=initial, where=where)
File ~/miniconda3/envs/np21/lib/python3.13/site-packages/numpy/_core/fromnumeric.py:84, in _wrapreduction(obj, ufunc, method, axis, dtype, out, **kwargs)
82 return reduction(axis=axis, dtype=dtype, out=out, **passkwargs)
83 else:
---> 84 return reduction(axis=axis, out=out, **passkwargs)
86 return ufunc.reduce(obj, axis, dtype, out, **passkwargs)
File ~/miniconda3/envs/np21/lib/python3.13/site-packages/numpy/ma/core.py:6091, in MaskedArray.max(self, axis, out, fill_value, keepdims)
6089 # Get rid of Infs
6090 if newmask.ndim:
-> 6091 np.copyto(result, result.fill_value, where=newmask)
6092 elif newmask:
6093 result = masked
TypeError: Cannot cast scalar from dtype('int64') to dtype('uint8') according to the rule 'same_kind'
```
</details>
**Anything else we need to know?**:
I have tried explicitly setting `fill_value` to zero, but the error still happens.
```python
import dask.array as da
xx = da.arange(0, 10, dtype='uint8')
xx = da.ma.masked_where(xx == 1, xx)
da.ma.set_fill_value(xx, 0)
xx.max().compute()
```
**Environment**:
- Dask version: 2024.11.2
- Python version: 3.13.0 | packaged by conda-forge | (main, Nov 27 2024, 19:18:26) [Clang 18.1.8 ]
- Operating System: macOS
- Install method (conda, pip, source): conda
- numpy: 2.1.3
|
closed
|
2024-12-03T04:40:34Z
|
2024-12-18T12:03:37Z
|
https://github.com/dask/dask/issues/11574
|
[
"needs triage"
] |
Kirill888
| 1
|
ClimbsRocks/auto_ml
|
scikit-learn
| 309
|
make sure compare_all_algos lives up to the name
|
and if so, remove duplicate tests for compare all models
|
open
|
2017-08-02T01:29:12Z
|
2017-08-02T01:29:12Z
|
https://github.com/ClimbsRocks/auto_ml/issues/309
|
[] |
ClimbsRocks
| 0
|
alteryx/featuretools
|
scikit-learn
| 2,115
|
Feature serialization and deserialization improvements
|
The serialization and deserialization of features should be improved.
For serialization we should update the approach so we are not duplicating primitive serialized primitive information. Currently if there are features that share the same primitive, each serialized feature contains the information needed to deserialize the primitive. This primitive information only needs to be stored once and not duplicated. We should separate primitive information from feature information and just link from the feature to the corresponding primitive information.
For deserialization we currently create separate instances of primitives for each feature. For features that share the same primitive (with the same arguments) this means we have separate, but identical instances of the primitive. We should update feature deserialization so that features that share a primitive reference the same primitive instance rather than creating unique instances.
|
closed
|
2022-06-16T21:27:11Z
|
2022-07-06T17:58:54Z
|
https://github.com/alteryx/featuretools/issues/2115
|
[] |
thehomebrewnerd
| 0
|
xlwings/xlwings
|
automation
| 2,135
|
Error exporting cmap formatted DF to Excel
|
#### OS (e.g. Windows 10 or macOS Sierra)
Windows 10
#### Versions of xlwings, Excel and Python (e.g. 0.11.8, Office 365, Python 3.7)
Running on Jupyter Notebook
#### Describe your issue (incl. Traceback!)
Hi, I am having an issue pasting dataframes that is formatted via background_gradient through pandas into Excel.
An unformatted dataframe works perfect but formatted ones throw up an error.
```python
# Your traceback here
TypeError Traceback (most recent call last)
TypeError: must be real number, not Styler
SystemError: <built-in method Invoke of PyIDispatch object at 0x0000013BAF0C11D8> returned a result with an error set
#### Include a minimal code sample to reproduce the issue (and attach a sample workbook if required!)
```python
import pandas as pd
import xlwings as xw
import numpy as np
df = pd.DataFrame(np.random.randint(0,100,size=(100, 4)), columns=list('ABCD'))
df
wb = xw.Book()
ws1 = wb.sheets['Sheet1']
ws1['B2'].options(index=False).value = df
# works well until here
df1 = df.style.background_gradient(axis = 1, cmap = 'RdYlGn')
df1
# error when pasting df1
Thank you.
```
|
closed
|
2023-01-06T07:38:38Z
|
2023-01-17T10:42:58Z
|
https://github.com/xlwings/xlwings/issues/2135
|
[] |
russwongg
| 5
|
slackapi/python-slack-sdk
|
asyncio
| 913
|
Updated pop up modal view seen for a second and closed immediately
|
### Description
Not sure what’s is missing , pasting the exact code.When I use below method I'm able to see pop getting update on click of submit button but its closing within approximately 2 seconds
```python
def send_views(user_name):
form_json = json.loads(request.form.get('payload'))
open_dialog1 = slack_client.api_call("views.update",
trigger_id=form_json[ "trigger_id" ],
view_id = form_json["view"]["id"],
hash=form_json["view"]["hash"],
view=views2
)
```
Below is views2 json
```json
views2 = {
"title": {
"type": "plain_text",
"text": "My App",
"emoji": True
},
"type": "modal",
"close": {
"type": "plain_text",
"text": "Cancel",
"emoji": True
},
"blocks": [
{
"type": "section",
"text": {
"type": "plain_text",
"text": "Submitted Successfully",
"emoji": True
}
}
]
}
```
Initial view
### What type of issue is this? (place an `x` in one of the `[ ]`)
- [ ] bug
- [ ] enhancement (feature request)
- [ x] question
- [ ] documentation related
- [ ] testing related
- [ ] discussion
### Requirements (place an `x` in each of the `[ ]`)
* [x ] I've read and understood the [Contributing guidelines](https://github.com/slackapi/node-slack-sdk/blob/master/.github/contributing.md) and have done my best effort to follow them.
* [x] I've read and agree to the [Code of Conduct](https://slackhq.github.io/code-of-conduct).
* [ x] I've searched for any related issues and avoided creating a duplicate issue.
---
### Bug Report
Filling out the following details about bugs will help us solve your issue sooner.
#### Packages:
Select all that apply:
- [ ] `@slack/web-api`
- [x] `@slack/events-api`
- [x] `@slack/interactive-messages`
- [ ] `@slack/rtm-api`
- [ ] `@slack/webhooks`
- [ ] `@slack/oauth`
- [ ] I don't know
#### Reproducible in:
package version:
node version:
OS version(s):
#### Steps to reproduce:
1.
2.
3.
#### Expected result:
I want to see initial view getting update to "view2"
#### Actual result:
Updated pop up only seen for split second on clicking submit button and closes
What actually happened
#### Attachments:
Logs, screenshots, screencast, sample project, funny gif, etc.
|
closed
|
2021-01-11T19:52:32Z
|
2021-01-14T15:11:42Z
|
https://github.com/slackapi/python-slack-sdk/issues/913
|
[
"question",
"web-client"
] |
ravi7271973
| 10
|
junyanz/pytorch-CycleGAN-and-pix2pix
|
deep-learning
| 1,052
|
WaterColor Style transfer, the generated image is not obvious...
|
Hello, I'm using cycleGAN to do watercolor style transfer, and I have collected many watercolor style images (about 3500) of different scenes with different objects. But the generated image is just no different of the original.
Some questions in my mind:
1) should the training set keep in the similar scene or similar shape of objects?
2) if the above 1 is correct, should I choose the training set with similar shape of objects (just like horse->zebra, and in my condition, I should choose real scene people -> watercolor painting people), and retrain or fine-tuning the model on these rechosen dataset?
3) If I keep the original dataset, what should I do to train the model to learn the watercolor style?
Some advices? Thank you so much.
|
open
|
2020-06-01T03:40:14Z
|
2020-06-01T06:01:50Z
|
https://github.com/junyanz/pytorch-CycleGAN-and-pix2pix/issues/1052
|
[] |
Rancherzhang
| 2
|
wkentaro/labelme
|
deep-learning
| 435
|
Feedback————How can I change the color of the 'label_viz' background?
|
Hello, wkentaro. I want to implement some display functions about my data, but I don't know how to get it.
I will get a folder in command line with " labelme_json_to_dataset /path ", this folder contains an image called 'label_viz'.

I try to change the color of label 'airplane' but I don't know how to change the background. And the background color is not completely black'0' but it seems to be blurred. How can I change it?
Thanks!!
|
closed
|
2019-07-03T01:31:28Z
|
2019-07-04T08:47:43Z
|
https://github.com/wkentaro/labelme/issues/435
|
[] |
DahuoJ
| 7
|
dmlc/gluon-cv
|
computer-vision
| 1,071
|
Using the provided script train.py gives error
|
Windows 8, Python 3.8, no GPU
I have looked online for solutions and tried:
```python
for param in self.net.collect_params().values():
if param._data is None:
param.initialize()
```
However, the error remains still. Please advise. Thanks for your help.
### Terminal output (Powershell) for running train.py
PS C:\Users\Nam\Desktop\elen 331 final project> **python train.py --dataset ade20k --model psp --backbone resnet50 --syncbn --epochs 120 --lr 0.01 --checkname mycheckpoint**
Number of GPUs: 0
Namespace(aux=False, aux_weight=0.5, backbone='resnet50', base_size=520, batch_size=16, checkname='mycheckpoint', crop_size=480, ctx=[], dataset='ade20k', dtype='float32', epochs=120, eval=False, kvstore='device', lr=0.01, model='psp', model_zoo=None, momentum=0.9, ngpus=0, no_cuda=False, no_val=False, no_wd=False, norm_kwargs={'num_devices': 0}, norm_layer=<class 'mxnet.gluon.contrib.nn.basic_layers.SyncBatchNorm'>, resume=None, start_epoch=0, syncbn=True, test_batch_size=16, train_split='train', weight_decay=0.0001, workers=16)
self.crop_size 480
PSPNet(
(conv1): HybridSequential(
(0): Conv2D(3 -> 64, kernel_size=(3, 3), stride=(2, 2), padding=(1, 1), bias=False)
(1): SyncBatchNorm(eps=1e-05, momentum=0.9, fix_gamma=False, use_global_stats=False, ndev=0, key='pspnet0_resnetv1s_syncbatchnorm0_', in_channels=64)
(2): Activation(relu)
(3): Conv2D(64 -> 64, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)
(4): SyncBatchNorm(eps=1e-05, momentum=0.9, fix_gamma=False, use_global_stats=False, ndev=0, key='pspnet0_resnetv1s_syncbatchnorm1_', in_channels=64)
(5): Activation(relu)
(6): Conv2D(64 -> 128, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)
)
(bn1): SyncBatchNorm(eps=1e-05, momentum=0.9, fix_gamma=False, use_global_stats=False, ndev=0, key='pspnet0_resnetv1s_syncbatchnorm2_', in_channels=128)
(relu): Activation(relu)
(maxpool): MaxPool2D(size=(3, 3), stride=(2, 2), padding=(1, 1), ceil_mode=False, global_pool=False, pool_type=max, layout=NCHW)
(layer1): HybridSequential(
(0): BottleneckV1b(
(conv1): Conv2D(128 -> 64, kernel_size=(1, 1), stride=(1, 1), bias=False)
(bn1): SyncBatchNorm(eps=1e-05, momentum=0.9, fix_gamma=False, use_global_stats=False, ndev=0, key='pspnet0_resnetv1s_layers1_syncbatchnorm0_', in_channels=64)
(relu1): Activation(relu)
(conv2): Conv2D(64 -> 64, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)
(bn2): SyncBatchNorm(eps=1e-05, momentum=0.9, fix_gamma=False, use_global_stats=False, ndev=0, key='pspnet0_resnetv1s_layers1_syncbatchnorm1_', in_channels=64)
(relu2): Activation(relu)
(conv3): Conv2D(64 -> 256, kernel_size=(1, 1), stride=(1, 1), bias=False)
(bn3): SyncBatchNorm(eps=1e-05, momentum=0.9, fix_gamma=False, use_global_stats=False, ndev=0, key='pspnet0_resnetv1s_layers1_syncbatchnorm2_', in_channels=256)
(relu3): Activation(relu)
(downsample): HybridSequential(
(0): Conv2D(128 -> 256, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): SyncBatchNorm(eps=1e-05, momentum=0.9, fix_gamma=False, use_global_stats=False, ndev=0, key='pspnet0_resnetv1s_down1_syncbatchnorm0_', in_channels=256)
)
)
(1): BottleneckV1b(
(conv1): Conv2D(256 -> 64, kernel_size=(1, 1), stride=(1, 1), bias=False)
(bn1): SyncBatchNorm(eps=1e-05, momentum=0.9, fix_gamma=False, use_global_stats=False, ndev=0, key='pspnet0_resnetv1s_layers1_syncbatchnorm3_', in_channels=64)
(relu1): Activation(relu)
(conv2): Conv2D(64 -> 64, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)
(bn2): SyncBatchNorm(eps=1e-05, momentum=0.9, fix_gamma=False, use_global_stats=False, ndev=0, key='pspnet0_resnetv1s_layers1_syncbatchnorm4_', in_channels=64)
(relu2): Activation(relu)
(conv3): Conv2D(64 -> 256, kernel_size=(1, 1), stride=(1, 1), bias=False)
(bn3): SyncBatchNorm(eps=1e-05, momentum=0.9, fix_gamma=False, use_global_stats=False, ndev=0, key='pspnet0_resnetv1s_layers1_syncbatchnorm5_', in_channels=256)
(relu3): Activation(relu)
)
(2): BottleneckV1b(
(conv1): Conv2D(256 -> 64, kernel_size=(1, 1), stride=(1, 1), bias=False)
(bn1): SyncBatchNorm(eps=1e-05, momentum=0.9, fix_gamma=False, use_global_stats=False, ndev=0, key='pspnet0_resnetv1s_layers1_syncbatchnorm6_', in_channels=64)
(relu1): Activation(relu)
(conv2): Conv2D(64 -> 64, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)
(bn2): SyncBatchNorm(eps=1e-05, momentum=0.9, fix_gamma=False, use_global_stats=False, ndev=0, key='pspnet0_resnetv1s_layers1_syncbatchnorm7_', in_channels=64)
(relu2): Activation(relu)
(conv3): Conv2D(64 -> 256, kernel_size=(1, 1), stride=(1, 1), bias=False)
(bn3): SyncBatchNorm(eps=1e-05, momentum=0.9, fix_gamma=False, use_global_stats=False, ndev=0, key='pspnet0_resnetv1s_layers1_syncbatchnorm8_', in_channels=256)
(relu3): Activation(relu)
)
)
(layer2): HybridSequential(
(0): BottleneckV1b(
(conv1): Conv2D(256 -> 128, kernel_size=(1, 1), stride=(1, 1), bias=False)
(bn1): SyncBatchNorm(eps=1e-05, momentum=0.9, fix_gamma=False, use_global_stats=False, ndev=0, key='pspnet0_resnetv1s_layers2_syncbatchnorm0_', in_channels=128)
(relu1): Activation(relu)
(conv2): Conv2D(128 -> 128, kernel_size=(3, 3), stride=(2, 2), padding=(1, 1), bias=False)
(bn2): SyncBatchNorm(eps=1e-05, momentum=0.9, fix_gamma=False, use_global_stats=False, ndev=0, key='pspnet0_resnetv1s_layers2_syncbatchnorm1_', in_channels=128)
(relu2): Activation(relu)
(conv3): Conv2D(128 -> 512, kernel_size=(1, 1), stride=(1, 1), bias=False)
(bn3): SyncBatchNorm(eps=1e-05, momentum=0.9, fix_gamma=False, use_global_stats=False, ndev=0, key='pspnet0_resnetv1s_layers2_syncbatchnorm2_', in_channels=512)
(relu3): Activation(relu)
(downsample): HybridSequential(
(0): Conv2D(256 -> 512, kernel_size=(1, 1), stride=(2, 2), bias=False)
(1): SyncBatchNorm(eps=1e-05, momentum=0.9, fix_gamma=False, use_global_stats=False, ndev=0, key='pspnet0_resnetv1s_down2_syncbatchnorm0_', in_channels=512)
)
)
(1): BottleneckV1b(
(conv1): Conv2D(512 -> 128, kernel_size=(1, 1), stride=(1, 1), bias=False)
(bn1): SyncBatchNorm(eps=1e-05, momentum=0.9, fix_gamma=False, use_global_stats=False, ndev=0, key='pspnet0_resnetv1s_layers2_syncbatchnorm3_', in_channels=128)
(relu1): Activation(relu)
(conv2): Conv2D(128 -> 128, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)
(bn2): SyncBatchNorm(eps=1e-05, momentum=0.9, fix_gamma=False, use_global_stats=False, ndev=0, key='pspnet0_resnetv1s_layers2_syncbatchnorm4_', in_channels=128)
(relu2): Activation(relu)
(conv3): Conv2D(128 -> 512, kernel_size=(1, 1), stride=(1, 1), bias=False)
(bn3): SyncBatchNorm(eps=1e-05, momentum=0.9, fix_gamma=False, use_global_stats=False, ndev=0, key='pspnet0_resnetv1s_layers2_syncbatchnorm5_', in_channels=512)
(relu3): Activation(relu)
)
(2): BottleneckV1b(
(conv1): Conv2D(512 -> 128, kernel_size=(1, 1), stride=(1, 1), bias=False)
(bn1): SyncBatchNorm(eps=1e-05, momentum=0.9, fix_gamma=False, use_global_stats=False, ndev=0, key='pspnet0_resnetv1s_layers2_syncbatchnorm6_', in_channels=128)
(relu1): Activation(relu)
(conv2): Conv2D(128 -> 128, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)
(bn2): SyncBatchNorm(eps=1e-05, momentum=0.9, fix_gamma=False, use_global_stats=False, ndev=0, key='pspnet0_resnetv1s_layers2_syncbatchnorm7_', in_channels=128)
(relu2): Activation(relu)
(conv3): Conv2D(128 -> 512, kernel_size=(1, 1), stride=(1, 1), bias=False)
(bn3): SyncBatchNorm(eps=1e-05, momentum=0.9, fix_gamma=False, use_global_stats=False, ndev=0, key='pspnet0_resnetv1s_layers2_syncbatchnorm8_', in_channels=512)
(relu3): Activation(relu)
)
(3): BottleneckV1b(
(conv1): Conv2D(512 -> 128, kernel_size=(1, 1), stride=(1, 1), bias=False)
(bn1): SyncBatchNorm(eps=1e-05, momentum=0.9, fix_gamma=False, use_global_stats=False, ndev=0, key='pspnet0_resnetv1s_layers2_syncbatchnorm9_', in_channels=128)
(relu1): Activation(relu)
(conv2): Conv2D(128 -> 128, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)
(bn2): SyncBatchNorm(eps=1e-05, momentum=0.9, fix_gamma=False, use_global_stats=False, ndev=0, key='pspnet0_resnetv1s_layers2_syncbatchnorm10_', in_channels=128)
(relu2): Activation(relu)
(conv3): Conv2D(128 -> 512, kernel_size=(1, 1), stride=(1, 1), bias=False)
(bn3): SyncBatchNorm(eps=1e-05, momentum=0.9, fix_gamma=False, use_global_stats=False, ndev=0, key='pspnet0_resnetv1s_layers2_syncbatchnorm11_', in_channels=512)
(relu3): Activation(relu)
)
)
(layer3): HybridSequential(
(0): BottleneckV1b(
(conv1): Conv2D(512 -> 256, kernel_size=(1, 1), stride=(1, 1), bias=False)
(bn1): SyncBatchNorm(eps=1e-05, momentum=0.9, fix_gamma=False, use_global_stats=False, ndev=0, key='pspnet0_resnetv1s_layers3_syncbatchnorm0_', in_channels=256)
(relu1): Activation(relu)
(conv2): Conv2D(256 -> 256, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)
(bn2): SyncBatchNorm(eps=1e-05, momentum=0.9, fix_gamma=False, use_global_stats=False, ndev=0, key='pspnet0_resnetv1s_layers3_syncbatchnorm1_', in_channels=256)
(relu2): Activation(relu)
(conv3): Conv2D(256 -> 1024, kernel_size=(1, 1), stride=(1, 1), bias=False)
(bn3): SyncBatchNorm(eps=1e-05, momentum=0.9, fix_gamma=False, use_global_stats=False, ndev=0, key='pspnet0_resnetv1s_layers3_syncbatchnorm2_', in_channels=1024)
(relu3): Activation(relu)
(downsample): HybridSequential(
(0): Conv2D(512 -> 1024, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): SyncBatchNorm(eps=1e-05, momentum=0.9, fix_gamma=False, use_global_stats=False, ndev=0, key='pspnet0_resnetv1s_down3_syncbatchnorm0_', in_channels=1024)
)
)
(1): BottleneckV1b(
(conv1): Conv2D(1024 -> 256, kernel_size=(1, 1), stride=(1, 1), bias=False)
(bn1): SyncBatchNorm(eps=1e-05, momentum=0.9, fix_gamma=False, use_global_stats=False, ndev=0, key='pspnet0_resnetv1s_layers3_syncbatchnorm3_', in_channels=256)
(relu1): Activation(relu)
(conv2): Conv2D(256 -> 256, kernel_size=(3, 3), stride=(1, 1), padding=(2, 2), dilation=(2, 2), bias=False)
(bn2): SyncBatchNorm(eps=1e-05, momentum=0.9, fix_gamma=False, use_global_stats=False, ndev=0, key='pspnet0_resnetv1s_layers3_syncbatchnorm4_', in_channels=256)
(relu2): Activation(relu)
(conv3): Conv2D(256 -> 1024, kernel_size=(1, 1), stride=(1, 1), bias=False)
(bn3): SyncBatchNorm(eps=1e-05, momentum=0.9, fix_gamma=False, use_global_stats=False, ndev=0, key='pspnet0_resnetv1s_layers3_syncbatchnorm5_', in_channels=1024)
(relu3): Activation(relu)
)
(2): BottleneckV1b(
(conv1): Conv2D(1024 -> 256, kernel_size=(1, 1), stride=(1, 1), bias=False)
(bn1): SyncBatchNorm(eps=1e-05, momentum=0.9, fix_gamma=False, use_global_stats=False, ndev=0, key='pspnet0_resnetv1s_layers3_syncbatchnorm6_', in_channels=256)
(relu1): Activation(relu)
(conv2): Conv2D(256 -> 256, kernel_size=(3, 3), stride=(1, 1), padding=(2, 2), dilation=(2, 2), bias=False)
(bn2): SyncBatchNorm(eps=1e-05, momentum=0.9, fix_gamma=False, use_global_stats=False, ndev=0, key='pspnet0_resnetv1s_layers3_syncbatchnorm7_', in_channels=256)
(relu2): Activation(relu)
(conv3): Conv2D(256 -> 1024, kernel_size=(1, 1), stride=(1, 1), bias=False)
(bn3): SyncBatchNorm(eps=1e-05, momentum=0.9, fix_gamma=False, use_global_stats=False, ndev=0, key='pspnet0_resnetv1s_layers3_syncbatchnorm8_', in_channels=1024)
(relu3): Activation(relu)
)
(3): BottleneckV1b(
(conv1): Conv2D(1024 -> 256, kernel_size=(1, 1), stride=(1, 1), bias=False)
(bn1): SyncBatchNorm(eps=1e-05, momentum=0.9, fix_gamma=False, use_global_stats=False, ndev=0, key='pspnet0_resnetv1s_layers3_syncbatchnorm9_', in_channels=256)
(relu1): Activation(relu)
(conv2): Conv2D(256 -> 256, kernel_size=(3, 3), stride=(1, 1), padding=(2, 2), dilation=(2, 2), bias=False)
(bn2): SyncBatchNorm(eps=1e-05, momentum=0.9, fix_gamma=False, use_global_stats=False, ndev=0, key='pspnet0_resnetv1s_layers3_syncbatchnorm10_', in_channels=256)
(relu2): Activation(relu)
(conv3): Conv2D(256 -> 1024, kernel_size=(1, 1), stride=(1, 1), bias=False)
(bn3): SyncBatchNorm(eps=1e-05, momentum=0.9, fix_gamma=False, use_global_stats=False, ndev=0, key='pspnet0_resnetv1s_layers3_syncbatchnorm11_', in_channels=1024)
(relu3): Activation(relu)
)
(4): BottleneckV1b(
(conv1): Conv2D(1024 -> 256, kernel_size=(1, 1), stride=(1, 1), bias=False)
(bn1): SyncBatchNorm(eps=1e-05, momentum=0.9, fix_gamma=False, use_global_stats=False, ndev=0, key='pspnet0_resnetv1s_layers3_syncbatchnorm12_', in_channels=256)
(relu1): Activation(relu)
(conv2): Conv2D(256 -> 256, kernel_size=(3, 3), stride=(1, 1), padding=(2, 2), dilation=(2, 2), bias=False)
(bn2): SyncBatchNorm(eps=1e-05, momentum=0.9, fix_gamma=False, use_global_stats=False, ndev=0, key='pspnet0_resnetv1s_layers3_syncbatchnorm13_', in_channels=256)
(relu2): Activation(relu)
(conv3): Conv2D(256 -> 1024, kernel_size=(1, 1), stride=(1, 1), bias=False)
(bn3): SyncBatchNorm(eps=1e-05, momentum=0.9, fix_gamma=False, use_global_stats=False, ndev=0, key='pspnet0_resnetv1s_layers3_syncbatchnorm14_', in_channels=1024)
(relu3): Activation(relu)
)
(5): BottleneckV1b(
(conv1): Conv2D(1024 -> 256, kernel_size=(1, 1), stride=(1, 1), bias=False)
(bn1): SyncBatchNorm(eps=1e-05, momentum=0.9, fix_gamma=False, use_global_stats=False, ndev=0, key='pspnet0_resnetv1s_layers3_syncbatchnorm15_', in_channels=256)
(relu1): Activation(relu)
(conv2): Conv2D(256 -> 256, kernel_size=(3, 3), stride=(1, 1), padding=(2, 2), dilation=(2, 2), bias=False)
(bn2): SyncBatchNorm(eps=1e-05, momentum=0.9, fix_gamma=False, use_global_stats=False, ndev=0, key='pspnet0_resnetv1s_layers3_syncbatchnorm16_', in_channels=256)
(relu2): Activation(relu)
(conv3): Conv2D(256 -> 1024, kernel_size=(1, 1), stride=(1, 1), bias=False)
(bn3): SyncBatchNorm(eps=1e-05, momentum=0.9, fix_gamma=False, use_global_stats=False, ndev=0, key='pspnet0_resnetv1s_layers3_syncbatchnorm17_', in_channels=1024)
(relu3): Activation(relu)
)
)
(layer4): HybridSequential(
(0): BottleneckV1b(
(conv1): Conv2D(1024 -> 512, kernel_size=(1, 1), stride=(1, 1), bias=False)
(bn1): SyncBatchNorm(eps=1e-05, momentum=0.9, fix_gamma=False, use_global_stats=False, ndev=0, key='pspnet0_resnetv1s_layers4_syncbatchnorm0_', in_channels=512)
(relu1): Activation(relu)
(conv2): Conv2D(512 -> 512, kernel_size=(3, 3), stride=(1, 1), padding=(2, 2), dilation=(2, 2), bias=False)
(bn2): SyncBatchNorm(eps=1e-05, momentum=0.9, fix_gamma=False, use_global_stats=False, ndev=0, key='pspnet0_resnetv1s_layers4_syncbatchnorm1_', in_channels=512)
(relu2): Activation(relu)
(conv3): Conv2D(512 -> 2048, kernel_size=(1, 1), stride=(1, 1), bias=False)
(bn3): SyncBatchNorm(eps=1e-05, momentum=0.9, fix_gamma=False, use_global_stats=False, ndev=0, key='pspnet0_resnetv1s_layers4_syncbatchnorm2_', in_channels=2048)
(relu3): Activation(relu)
(downsample): HybridSequential(
(0): Conv2D(1024 -> 2048, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): SyncBatchNorm(eps=1e-05, momentum=0.9, fix_gamma=False, use_global_stats=False, ndev=0, key='pspnet0_resnetv1s_down4_syncbatchnorm0_', in_channels=2048)
)
)
(1): BottleneckV1b(
(conv1): Conv2D(2048 -> 512, kernel_size=(1, 1), stride=(1, 1), bias=False)
(bn1): SyncBatchNorm(eps=1e-05, momentum=0.9, fix_gamma=False, use_global_stats=False, ndev=0, key='pspnet0_resnetv1s_layers4_syncbatchnorm3_', in_channels=512)
(relu1): Activation(relu)
(conv2): Conv2D(512 -> 512, kernel_size=(3, 3), stride=(1, 1), padding=(4, 4), dilation=(4, 4), bias=False)
(bn2): SyncBatchNorm(eps=1e-05, momentum=0.9, fix_gamma=False, use_global_stats=False, ndev=0, key='pspnet0_resnetv1s_layers4_syncbatchnorm4_', in_channels=512)
(relu2): Activation(relu)
(conv3): Conv2D(512 -> 2048, kernel_size=(1, 1), stride=(1, 1), bias=False)
(bn3): SyncBatchNorm(eps=1e-05, momentum=0.9, fix_gamma=False, use_global_stats=False, ndev=0, key='pspnet0_resnetv1s_layers4_syncbatchnorm5_', in_channels=2048)
(relu3): Activation(relu)
)
(2): BottleneckV1b(
(conv1): Conv2D(2048 -> 512, kernel_size=(1, 1), stride=(1, 1), bias=False)
(bn1): SyncBatchNorm(eps=1e-05, momentum=0.9, fix_gamma=False, use_global_stats=False, ndev=0, key='pspnet0_resnetv1s_layers4_syncbatchnorm6_', in_channels=512)
(relu1): Activation(relu)
(conv2): Conv2D(512 -> 512, kernel_size=(3, 3), stride=(1, 1), padding=(4, 4), dilation=(4, 4), bias=False)
(bn2): SyncBatchNorm(eps=1e-05, momentum=0.9, fix_gamma=False, use_global_stats=False, ndev=0, key='pspnet0_resnetv1s_layers4_syncbatchnorm7_', in_channels=512)
(relu2): Activation(relu)
(conv3): Conv2D(512 -> 2048, kernel_size=(1, 1), stride=(1, 1), bias=False)
(bn3): SyncBatchNorm(eps=1e-05, momentum=0.9, fix_gamma=False, use_global_stats=False, ndev=0, key='pspnet0_resnetv1s_layers4_syncbatchnorm8_', in_channels=2048)
(relu3): Activation(relu)
)
)
(head): _PSPHead(
(psp): _PyramidPooling(
(conv1): HybridSequential(
(0): Conv2D(2048 -> 512, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): SyncBatchNorm(eps=1e-05, momentum=0.9, fix_gamma=False, use_global_stats=False, ndev=0, key='pspnet0__pyramidpooling0_hybridsequential0_syncbatchnorm0_', in_channels=512)
(2): Activation(relu)
)
(conv2): HybridSequential(
(0): Conv2D(2048 -> 512, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): SyncBatchNorm(eps=1e-05, momentum=0.9, fix_gamma=False, use_global_stats=False, ndev=0, key='pspnet0__pyramidpooling0_hybridsequential1_syncbatchnorm0_', in_channels=512)
(2): Activation(relu)
)
(conv3): HybridSequential(
(0): Conv2D(2048 -> 512, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): SyncBatchNorm(eps=1e-05, momentum=0.9, fix_gamma=False, use_global_stats=False, ndev=0, key='pspnet0__pyramidpooling0_hybridsequential2_syncbatchnorm0_', in_channels=512)
(2): Activation(relu)
)
(conv4): HybridSequential(
(0): Conv2D(2048 -> 512, kernel_size=(1, 1), stride=(1, 1), bias=False)
(1): SyncBatchNorm(eps=1e-05, momentum=0.9, fix_gamma=False, use_global_stats=False, ndev=0, key='pspnet0__pyramidpooling0_hybridsequential3_syncbatchnorm0_', in_channels=512)
(2): Activation(relu)
)
)
(block): HybridSequential(
(0): Conv2D(4096 -> 512, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1), bias=False)
(1): SyncBatchNorm(eps=1e-05, momentum=0.9, fix_gamma=False, use_global_stats=False, ndev=0, key='pspnet0__psphead0_syncbatchnorm0_', in_channels=512)
(2): Activation(relu)
(3): Dropout(p = 0.1, axes=())
(4): Conv2D(512 -> 150, kernel_size=(1, 1), stride=(1, 1))
)
)
)
C:\Users\Nam\AppData\Local\Programs\Python\Python37\lib\site-packages\mxnet\gluon\parameter.py:862: UserWarning: Parameter 'pspnet0_resnetv1s_conv0_weight' is already initialized, ignoring. Set force_reinit=True to re-initialize.
v.initialize(None, ctx, init, force_reinit=force_reinit)
Traceback (most recent call last):
File "train.py", line 224, in <module>
trainer = Trainer(args)
File "train.py", line 138, in __init__
self.evaluator = DataParallelModel(SegEvalModel(model), args.ctx)
File "C:\Users\Nam\AppData\Local\Programs\Python\Python37\lib\site-packages\gluoncv\utils\parallel.py", line 170, in __init__
module.collect_params().reset_ctx(ctx=ctx_list)
File "C:\Users\Nam\AppData\Local\Programs\Python\Python37\lib\site-packages\mxnet\gluon\parameter.py", line 879, in reset_ctx
i.reset_ctx(ctx)
File "C:\Users\Nam\AppData\Local\Programs\Python\Python37\lib\site-packages\mxnet\gluon\parameter.py", line 464, in reset_ctx
"has not been initialized."%self.name)
**ValueError: Cannot reset context for Parameter 'pspnet0_resnetv1s_conv0_weight' because it has not been initialized.**
|
closed
|
2019-12-02T21:53:08Z
|
2020-02-04T19:28:33Z
|
https://github.com/dmlc/gluon-cv/issues/1071
|
[] |
NamTran838P
| 11
|
d2l-ai/d2l-en
|
deep-learning
| 2,440
|
Wrong epanechikov kernel
|
Chapter 11.2. It is triangular kernel.
|
open
|
2023-02-12T15:50:31Z
|
2023-02-12T15:50:31Z
|
https://github.com/d2l-ai/d2l-en/issues/2440
|
[] |
yongduek
| 0
|
jofpin/trape
|
flask
| 376
|
Hey
|
closed
|
2023-03-11T18:34:19Z
|
2023-03-11T18:46:06Z
|
https://github.com/jofpin/trape/issues/376
|
[] |
Baggy79
| 1
|
|
seleniumbase/SeleniumBase
|
pytest
| 2,910
|
Add advanced UC Mode PyAutoGUI clicking methods
|
## Add advanced UC Mode PyAutoGUI clicking methods
```python
driver.uc_gui_click_x_y(x, y, timeframe=0.25) # PyAutoGUI click screen
driver.uc_gui_click_cf(frame="iframe", retry=False, blind=False) # (*)
```
👤 `driver.uc_gui_click_x_y(x, y, timeframe=0.25)` uses `PyAutoGUI` to click the screen at the coordinates. The mouse movement will take the duration of the timeframe.
👤 `driver.uc_gui_click_cf(frame="iframe", retry=False, blind=False)` has three args. (All optional). The first one, `frame`, lets you specify the iframe in case the CAPTCHA is not located in the first iframe on the page. The second one, `retry`, lets you retry the click after reloading the page if the first one didn't work (and a CAPTCHA is still present after the page reload). The third arg, `blind`, will retry after a page reload (if the first click failed) by clicking at the last known coordinates of the CAPTCHA checkbox without confirming first with Selenium that a CAPTCHA is still on the page.
|
closed
|
2024-07-06T06:24:50Z
|
2024-07-08T21:24:39Z
|
https://github.com/seleniumbase/SeleniumBase/issues/2910
|
[
"enhancement",
"UC Mode / CDP Mode"
] |
mdmintz
| 2
|
voila-dashboards/voila
|
jupyter
| 1,486
|
Broken rendering of math/LaTeX using HTMLMath widget since 0.5.5
|
Hello!
First of all, thanks for this tool it is awesome.
## Description
`HTMLMath` widget has not rendered Math/LaTeX content correctly since `0.5.5`.
<img width="960" alt="repro-0 5 5-bad" src="https://github.com/user-attachments/assets/eef21090-88ab-48b7-9b3d-94f8cacc52fb">
Going back to `0.5.4` it renders OK.
<img width="960" alt="repro-0 5 4-ok" src="https://github.com/user-attachments/assets/9e2d5b02-a15d-497d-b7ed-66bc4bbae3b6">
From the changelog, #1410 might have introduced the issue.
## Reproduce
Create a notebook with the following content (or download [repro.ipynb](https://github.com/akielbowicz/ipywidgets-playground/blob/voila-issue/voila-issue/repro.ipynb
))
```python
from ipywidgets import HTMLMath
HTMLMath("Some text with math $E = m c^2$")
```
```sh
python -m venv .venv
# activate venv
python -m pip install voila==0.5.5 ipywidgets jupyterlab
voila repro.ipynb
```
## Expected behavior
Math/LaTeX content of HTMLMath widget renders correctly.
## Context
- voila version 0.5.5, 0.5.6, 0.5.7
- Operating System and version: Tested on Windows 10/11 and within a docker container
- Browser and version: Edge, Librewolf, Brave
|
closed
|
2024-08-01T23:09:37Z
|
2024-10-09T20:54:11Z
|
https://github.com/voila-dashboards/voila/issues/1486
|
[
"bug"
] |
akielbowicz
| 1
|
piskvorky/gensim
|
nlp
| 2,728
|
[QUESTION] Update LDA model
|
## Problem description
Update an existing LDA model with an incremental approach. We create a LDA model for a collection of documents on demand basis. We save the resulting model file on the cloud. When a new LDA request arrives with fresh data, I need a way to incrementally update the model (live training) wit the these data. Typically I would use `lda.update()`. But what happens when the `lda.update()` takes as input a corpus that includes the same documents of the previous model?
Assumed to have `model1` trained on `corpus1` and a new `corpus2`, which is the best approach to do the incremental training of the new `model2` against `corpus2`?
I have seen a `lda.diff` function. So one could train `model2` and then run `mdiff, annotation = model1.diff(model2)`, then check `diff` and `annotation` and decide to promote `model2`. Does it make sense? Which is the best criterion to promote the new model then?
Thank you in advance!
#### Steps/code/corpus to reproduce
```python
from smart_open import open
# load the existing LDA model
current_model = LdaModel.load(open(s3_model_path))
# load the corpus
data = TextCorpus( open(s3_corpus_path) )
corpus_sentences = data.get_texts()
dictionary = Dictionary(corpus_sentences)
corpus = [dictionary.doc2bow(text) for text in corpus_sentences]
# update current model on the corpus
current_model.update(corpus)
```
#### Versions
```
Linux-5.0.0-36-generic-x86_64-with-Ubuntu-18.04-bionic
Python 3.6.9 (default, Nov 7 2019, 10:44:02)
[GCC 8.3.0]
NumPy 1.17.4
SciPy 1.0.0
gensim 3.8.1
FAST_VERSION 1
```
|
closed
|
2020-01-16T13:23:23Z
|
2020-01-23T07:40:12Z
|
https://github.com/piskvorky/gensim/issues/2728
|
[] |
loretoparisi
| 1
|
samuelcolvin/dirty-equals
|
pytest
| 38
|
Include documentation on how this works
|
I find this functionality quite interesting and think it could be useful.
But I'm a bit hesitant to include a library with "magical" behavior that I don't understand.
My main question is, why is the `__eq__` method of the right operand used?
The [python documentation](https://docs.python.org/3/reference/datamodel.html#object.__eq__) seems to suggest that `x == y` should call `x.__eq__(y)`.
> If the operands are of different types, and right operand’s type is a direct or indirect subclass of the left operand’s type, the reflected method of the right operand has priority, otherwise the left operand’s method has priority.
The last paragraph in the docs above point me to the `__eq__` implementation in the `DirtyEqualsMeta` class. But I'm not sure what is going on.
|
closed
|
2022-05-09T11:04:01Z
|
2022-05-13T14:01:32Z
|
https://github.com/samuelcolvin/dirty-equals/issues/38
|
[] |
Marco-Kaulea
| 3
|
strawberry-graphql/strawberry-django
|
graphql
| 712
|
Cannot find 'assumed_related_name' on 'YourRelatedModel' object, 'assumed_related_name' is an invalid parameter to prefetch_related()
|
## The Bug
When defining models in django, it is often assumed that we provide the `related_name` field. Usually, it is a plural of the name of the model we are pointing from. In `strawberry-django`, if this plural name is not provided for `related_name`, we are thrown the error:
```shell
Cannot find 'transactions' on Document object, 'transactions' is an invalid parameter to prefetch_related()
```
In other words—regardless of what my `related_name` actually is, `'transactions'` is being passed in.
For ex:
```python
class Document(Model):
name = CharField()
class Transaction(Model):
name = CharField()
document = ForeignKey(
"Document",
related_name="something_other_than_transactions", # this would work if "transactions" was passed in,
)
```
Note that this error is still present even if the query does not include the field, _and also_ if the object does not even declare the related type.
## Why this is an issue
I have a model that has two foreign keys to the same table. In order to make this framework work as expected, I have to change my core data models—not something I can really afford to do for performance reasons.
Lmk if there's anything other context I can provide, I think this ought to be enough.
|
closed
|
2025-02-24T07:49:40Z
|
2025-02-24T08:10:22Z
|
https://github.com/strawberry-graphql/strawberry-django/issues/712
|
[
"bug"
] |
jamietdavidson
| 1
|
frappe/frappe
|
rest-api
| 31,155
|
Asset Image can't be changed/removed once uploaded and Saved. Draft/Submitted
|
**Description:** The Image Field in the Asset is an "Allow on Submit" Field, allowing users to Add/Change the Image after Submitting. But once a User uploads an Image, it can't be removed or changed even though the option is there, it is not working.
**Pre-Requisites:** Create an Asset(Either Draft or Submitted).
**Steps to Recreate the Bug:**
1. Click "Change" (in front of the Image Area in Asset).
2. Click "Upload" and Select an Image.
3. Click "Save".
4. Click on the Image and click "Remove". Or "Upload" a different image.
5. Click "Save"
**ERPNext:** v14.74.4 (version-14)
**Frappe Framework:** v14.82.1 (version-14)
[Click here for the Screen Recording](https://youtu.be/jQu1bAzVx_k)
|
open
|
2025-02-06T05:10:53Z
|
2025-02-06T05:11:41Z
|
https://github.com/frappe/frappe/issues/31155
|
[
"bug"
] |
ransikerandeni
| 0
|
ghtmtt/DataPlotly
|
plotly
| 33
|
Animations are available for offline
|
A bit complicated to implement, but the code is more or less straightforward:
```python
f1 = [125.7, 219.12, 298.55, 132.32, 520.6, 3435.49, 2322.61, 1891.63, 216.97, 383.98, 82.01, 365.56, 199.98, 308.71, 217.58, 436.09, 711.77]
f2 = [1046.67, 1315.0, 1418.0, 997.33, 2972.3, 9700.0, 6726.0, 6002.5, 2096.0, 2470.0, 867.0, 2201.7, 1685.6, 2416.7, 1618.3, 2410.0, 2962.0]
trace1 = go.Scatter(
x = f1,
y = f2,
mode = 'markers'
)
layout = go.Layout(
xaxis = dict(range=[min(f1), max(f1)], autorange=False),
yaxis = dict(range=[min(f2), max(f2)], autorange=False),
updatemenus = [{'type': 'buttons', 'buttons':
[{'label': 'Play',
'method': 'animate',
'args': [None]},
{'label': 'Pause',
'method': 'animate',
'args': [[None],
{'frame' : {'duration':0, 'redraw': False}, 'mode':'immediate'},
]}
]}],
)
frames= []
for i, j in zip(f1, f2):
frames.append({'data': [{'x': [i], 'y': [j]}]})
data = [trace1]
fig = go.Figure(data=data, layout = layout, frames=frames)
plotly.offline.plot(fig)
```
API here
https://plot.ly/python/animations/#moving-frenet-frame-along-a-planar-curve
|
open
|
2017-07-07T13:05:38Z
|
2021-08-26T05:56:17Z
|
https://github.com/ghtmtt/DataPlotly/issues/33
|
[
"enhancement"
] |
ghtmtt
| 5
|
Python3WebSpider/ProxyPool
|
flask
| 18
|
zincrby 新的版本redis-py有改动
|
zincrby(name, amount, value)
需要将源代码中的zincrby第二、三参数换个顺序
|
closed
|
2018-12-24T12:47:51Z
|
2020-02-19T16:57:53Z
|
https://github.com/Python3WebSpider/ProxyPool/issues/18
|
[] |
duolk
| 1
|
graphql-python/gql
|
graphql
| 188
|
Customizable ExecutionResult, or how to get other API response parts
|
https://github.com/graphql-python/gql/blob/459d5ebacce95c817effcd18be152d6cf360cf2b/gql/transport/requests.py#L174
### Case:
The GraphQL API that I need to make requests to, responds with some other useful information, except of `data` block:
```json
{
"data": {...},
"extensions": {
"pageInfo": {
"limitPage": 200,
"totalCount": 23306516,
"hasNextPage": true,
"lastId": 41922710
}
}
}
```
But, as **gql** transport composes `ExecutionResult` from `data` and `errors` blocks only, I'm not able to get and use the `extensions` part in my code.
I was hoping to use `hasNextPage` and `lastId` values to iterate through pages of data, to load all the data to database.
### Question:
Is there any way to get `extensions` part of response, along with `data` and `errors`, using **gql**?
|
closed
|
2021-01-23T10:45:35Z
|
2021-11-24T09:10:03Z
|
https://github.com/graphql-python/gql/issues/188
|
[
"type: feature"
] |
extrin
| 7
|
ymcui/Chinese-LLaMA-Alpaca
|
nlp
| 295
|
合并词表报错
|
合并词表时我用了redpajama的tokenizer替换了llama的tokenizer,报错:
<img width="882" alt="image" src="https://github.com/ymcui/Chinese-LLaMA-Alpaca/assets/69674181/e4547f0e-f57d-4a6d-b20c-dc5436483d9d">
请问如何解决这种报错、或者说有什么解决的思路吗
|
closed
|
2023-05-10T06:55:26Z
|
2023-05-20T22:02:04Z
|
https://github.com/ymcui/Chinese-LLaMA-Alpaca/issues/295
|
[
"stale"
] |
han508
| 5
|
sktime/sktime
|
scikit-learn
| 7,950
|
[BUG] Link to documentation in ExpandingGreedySplitter display goes to incorrect URL
|
**Describe the bug**
The image displayed with you instantiate an `ExpandingGreedySplitter` object has a `?` icon in the top right that links to the API reference for the object. However rather than link to the `latest` or `stable` page, it links to the version of `sktime` that you have installed. In my case, it is `v0.36.0`, which displays "404 Documentation page not found".
i.e. `https://www.sktime.net/en/v0.36.0/api_reference/auto_generated/sktime.split.expandinggreedy.ExpandingGreedySplitter.html`
**To Reproduce**
```python
from sktime.split import ExpandingGreedySplitter
ExpandingGreedySplitter(12)
```
**Expected behavior**
The URL should link a page that works. Either `https://www.sktime.net/en/latest/api_reference/auto_generated/sktime.split.ExpandingGreedySplitter.html` or `https://www.sktime.net/en/stable/api_reference/auto_generated/sktime.split.ExpandingGreedySplitter.html`. I do not know whether `latest` or `stable` is preferred.
**Additional context**
<!--
Add any other context about the problem here.
-->
**Versions**
<details>
<!--
Please run the following code snippet and paste the output here:
from sktime import show_versions; show_versions()
-->
```python
Python dependencies:
pip: 25.0
sktime: 0.36.0
sklearn: 1.6.1
skbase: 0.12.0
numpy: 2.0.1
scipy: 1.15.1
pandas: 2.2.3
matplotlib: 3.10.0
joblib: 1.4.2
numba: None
statsmodels: 0.14.4
pmdarima: 2.0.4
statsforecast: None
tsfresh: None
tslearn: None
torch: None
tensorflow: None
```
</details>
|
open
|
2025-03-07T17:55:13Z
|
2025-03-22T14:23:35Z
|
https://github.com/sktime/sktime/issues/7950
|
[
"bug",
"documentation"
] |
gbilleyPeco
| 5
|
feature-engine/feature_engine
|
scikit-learn
| 446
|
[ENHANCEMENT] change pandas.corr() by numpy alternative in correlation selectors
|
**Is your feature request related to a problem? Please describe.**
For the following feature selectors:
- [SmartCorrelatedSelection](https://feature-engine.readthedocs.io/en/1.3.x/api_doc/selection/SmartCorrelatedSelection.html)
- [DropCorrelatedFeatures](https://feature-engine.readthedocs.io/en/1.3.x/api_doc/selection/DropCorrelatedFeatures.html)
`pandas.corr(method='pearson')` is currently being used to calculate the correlation matrix. This is very slow in performance compared to using the `numpy` equivalent method.
An example of where `pandas.corr(method='pearson')` is used can be found [here](https://github.com/feature-engine/feature_engine/blob/1b558821a58552bfb1addf96a86d04302ff619c2/feature_engine/selection/smart_correlation_selection.py#L239).
**Describe the solution you'd like**
Refer to here for specific code change: [smart_correlation_selection.py](https://github.com/feature-engine/feature_engine/blob/1b558821a58552bfb1addf96a86d04302ff619c2/feature_engine/selection/smart_correlation_selection.py#L239)
Instead of this:
```python
_correlated_matrix = X[self.variables_].corr(method=self.method)
```
We can do this instead:
```python
if self.method == 'pearson':
_correlated_matrix = pd.DataFrame(np.corrcoef(X[self.variables_], rowvar=False), columns=X[self.variables_].columns)
_correlated_matrix.index = _correlated_matrix.columns
```
**Additional context**
I currently haven't looked into how to make the other methods faster (e.g.` method=spearman`), but this can also be considered in this feature.
|
closed
|
2022-05-08T04:28:21Z
|
2024-02-18T17:24:28Z
|
https://github.com/feature-engine/feature_engine/issues/446
|
[
"enhancement"
] |
lauradang
| 1
|
recommenders-team/recommenders
|
machine-learning
| 1,562
|
[BUG] ValueError: numpy.ndarray size changed, may indicate binary incompatibility. Expected 88 from C header, got 80 from PyObject
|
### The benchmark notebook got a Numpy error while running, like this:
<!--- Describe your issue/bug/request in detail -->
```
ValueError: numpy.ndarray size changed, may indicate binary incompatibility. Expected 88 from C header, got 80 from PyObject
```
I searched from the stackoverflow, it seems that I got the conflicted version of Numpy. I just go with the SETUP guide.
|
closed
|
2021-11-01T12:17:39Z
|
2021-11-02T19:54:44Z
|
https://github.com/recommenders-team/recommenders/issues/1562
|
[
"bug"
] |
mokeeqian
| 1
|
qwj/python-proxy
|
asyncio
| 101
|
api_* tests require an internet connection
|
It would be helpful for distro packaging if the api_* tests had a non-internet tunnel endpoint, such as a dedicated test local service which could be stood up for the purpose of testing, e.g. `httpbin`, which is available as a pytest plugin `pytest-httpbin`
|
open
|
2020-12-07T18:39:50Z
|
2020-12-07T18:39:50Z
|
https://github.com/qwj/python-proxy/issues/101
|
[] |
jayvdb
| 0
|
pytest-dev/pytest-cov
|
pytest
| 456
|
.coveragerc sometimes ignored
|
# Summary
In jaraco/skeleton, I've configured my projects to [ignore .tox](https://github.com/jaraco/skeleton/blob/86efb884f805a9e1f64661ec758f3bd084fed515/.coveragerc#L2) for coverage. Despite this setting, I often see `.tox` in the coverage results. ([example](https://github.com/jaraco/cssutils/runs/2082642025?check_suite_focus=true#step:5:120)).
Not all projects are affected. Some show a report without tox (using the same config).
Note the tox lines do appear to implicate black or virtualenv (especially as 'click' is mentioned).
## Expected vs actual result
Expect `.tox` to be omitted as indicated.
# Reproducer
I've tried reproducing the issue in a [minimal repo](/jaraco/pytest-cov-456), and I can get the `.tox` results to appear if I run tests on a simple repo with only `pytest-cov` and `pytest-black` installed and enabled. But in that case, when I add the .coveragerc file, `.tox` disappears from the report, as expected. So there's something else about the projects that's causing the `.coveragerc` to be ignored. ~~Note also that omitting `pytest-black` causes the `.tox` lines to be omitted.~~
## Versions
See example above for one configuration (using latest version of everything), but it affects lots of versions and libraries.
## Config
See [jaraco/cssutils](/jaraco/cssutils) for one affected project.
Any advice on how I can troubleshoot further, to narrow the cause?
|
closed
|
2021-03-11T02:58:38Z
|
2021-10-03T23:58:05Z
|
https://github.com/pytest-dev/pytest-cov/issues/456
|
[] |
jaraco
| 4
|
MycroftAI/mycroft-core
|
nlp
| 2,651
|
Custom wake up word is only detected once during the "start-mycroft.sh debug" and not detected at all after that
|
Hi Team,
I have trained my custom wake up word "mokato" using precise models. When I test it with "precise-listen -b mokato.net" I see many ! (positive detection) and the output of "precise-test mokato.net mokato" is also good (100% postive), I am pasting all info below. Now when I start the "start-mycroft.sh debug" it detect the custom wake up word only once while starting. Please help me I trained with some 342 audios.
Here is the output of "mycroft-config edit user"
{
"max_allowed_core_version": 20.2,
"listener": {
"wake_word": "mokato"
},
"hotwords": {
"mokato": {
"module": "precise",
"local_model_file": "/home/pumo/voice/mycroft-core/customwakup/mycroft-precise/mokato.pb"
}
}
}
**OutPut of precise-test mokato.net mokato :**
instructions for updating:
Please use `rate` instead of `keep_prob`. Rate should be set to `rate = 1 - keep_prob`.
Data: <TrainData wake_words=342 not_wake_words=0 test_wake_words=4 test_not_wake_words=0>
=== False Positives ===
=== False Negatives ===
=== Counts ===
False Positives: 0
True Negatives: 0
False Negatives: 0
True Positives: 4
=== Summary ===
4 out of 4
100.00%
0.00% false positives
0.00% false negatives
**Output from /var/log/mycroft/voice.log :**
2020-08-08 00:01:02.217 | INFO | 28768 | mycroft.client.speech.listener:create_wake_word_recognizer:323 | Creating wake word engine
2020-08-08 00:01:02.217 | INFO | 28768 | mycroft.client.speech.listener:create_wake_word_recognizer:346 | Using hotword entry for mokato
2020-08-08 00:01:02.218 | WARNING | 28768 | mycroft.client.speech.listener:create_wake_word_recognizer:348 | Phonemes are missing falling back to listeners configuration
2020-08-08 00:01:02.218 | WARNING | 28768 | mycroft.client.speech.listener:create_wake_word_recognizer:352 | Threshold is missing falling back to listeners configuration
2020-08-08 00:01:02.219 | INFO | 28768 | mycroft.client.speech.hotword_factory:load_module:386 | Loading "mokato" wake word via precise
2020-08-08 00:01:04.985 | INFO | 28768 | mycroft.client.speech.listener:create_wakeup_recognizer:360 | creating stand up word engine
2020-08-08 00:01:04.986 | INFO | 28768 | mycroft.client.speech.hotword_factory:load_module:386 | Loading "wake up" wake word via pocketsphinx
2020-08-08 00:01:05.006 | INFO | 28768 | mycroft.messagebus.client.client:on_open:67 | Connected
2020-08-08 00:01:07.473 | INFO | 28768 | mycroft.session:get:72 | New Session Start: cd65c37c-408b-47ca-bc7c-3b6b08b74b09
2020-08-08 00:01:07.475 | INFO | 28768 | __main__:handle_wakeword:67 | Wakeword Detected: mokato
2020-08-08 00:01:07.886 | INFO | 28768 | __main__:handle_record_begin:37 | Begin Recording...
2020-08-08 00:01:10.976 | INFO | 28768 | __main__:handle_record_end:45 | End Recording...
2020-08-08 00:01:15.904 | ERROR | 28768 | mycroft.client.speech.listener:transcribe:239 | list index out of range
2020-08-08 00:01:15.910 | ERROR | 28768 | mycroft.client.speech.listener:transcribe:240 | Speech Recognition could not understand audio
|
closed
|
2020-08-07T18:41:14Z
|
2020-12-09T05:42:07Z
|
https://github.com/MycroftAI/mycroft-core/issues/2651
|
[] |
abhisek-mishra
| 4
|
zihangdai/xlnet
|
tensorflow
| 274
|
What is the function of _sample_mask method?
|

It seems XLNet do not have MASK.
|
closed
|
2020-09-29T09:55:44Z
|
2020-09-29T12:06:19Z
|
https://github.com/zihangdai/xlnet/issues/274
|
[] |
guotong1988
| 1
|
mars-project/mars
|
pandas
| 3,176
|
[BUG] session creation error
|
<!--
Thank you for your contribution!
Please review https://github.com/mars-project/mars/blob/master/CONTRIBUTING.rst before opening an issue.
-->
**Describe the bug**
session creation error
**To Reproduce**
To help us reproducing this bug, please provide information below:
1. Python 3.7.13
2. Mars 9.0
3. //
4. Full stack of the error.
```
ERROR:mars.services.cluster.uploader:Failed to upload node info
Traceback (most recent call last):
File "D:\Compiler\Anaconda\envs\py37\lib\site-packages\mars\services\cluster\uploader.py", line 122, in upload_node_info
self._info.env = await asyncio.to_thread(gather_node_env)
File "D:\Compiler\Anaconda\envs\py37\lib\site-packages\mars\lib\aio\_threads.py", line 36, in to_thread
return await loop.run_in_executor(None, func_call)
File "D:\Compiler\Anaconda\envs\py37\lib\concurrent\futures\thread.py", line 57, in run
result = self.fn(*self.args, **self.kwargs)
File "D:\Compiler\Anaconda\envs\py37\lib\site-packages\mars\services\cluster\gather.py", line 75, in gather_node_env
cuda_info = mars_resource.cuda_info()
File "D:\Compiler\Anaconda\envs\py37\lib\site-packages\mars\resource.py", line 360, in cuda_info
products=[nvutils.get_device_info(idx).name for idx in range(gpu_count)],
File "D:\Compiler\Anaconda\envs\py37\lib\site-packages\mars\resource.py", line 360, in <listcomp>
products=[nvutils.get_device_info(idx).name for idx in range(gpu_count)],
File "D:\Compiler\Anaconda\envs\py37\lib\site-packages\mars\lib\nvutils.py", line 343, in get_device_info
uuid=uuid.UUID(bytes=uuid_t.bytes),
File "D:\Compiler\Anaconda\envs\py37\lib\uuid.py", line 169, in __init__
raise ValueError('bytes is not a 16-char string')
ValueError: bytes is not a 16-char string
ERROR:mars.services.cluster.uploader:Failed to upload node info: bytes is not a 16-char string
`---------------------------------------------------------------------------
ValueError Traceback (most recent call last)
~\AppData\Local\Temp\ipykernel_39180\3232464671.py in <module>
----> 1 mars.new_session()
D:\Compiler\Anaconda\envs\py37\lib\site-packages\mars\deploy\oscar\session.py in new_session(address, session_id, backend, default, new, **kwargs)
2038
2039 session = SyncSession.init(
-> 2040 address, session_id=session_id, backend=backend, new=new, **kwargs
2041 )
2042 if default:
D:\Compiler\Anaconda\envs\py37\lib\site-packages\mars\deploy\oscar\session.py in init(cls, address, session_id, backend, new, **kwargs)
1634 coro = _IsolatedSession.init(address, session_id, backend, new=new, **kwargs)
1635 fut = asyncio.run_coroutine_threadsafe(coro, isolation.loop)
-> 1636 isolated_session = fut.result()
1637 return SyncSession(address, session_id, isolated_session, isolation)
1638
D:\Compiler\Anaconda\envs\py37\lib\concurrent\futures\_base.py in result(self, timeout)
433 raise CancelledError()
434 elif self._state == FINISHED:
--> 435 return self.__get_result()
436 else:
437 raise TimeoutError()
D:\Compiler\Anaconda\envs\py37\lib\concurrent\futures\_base.py in __get_result(self)
382 def __get_result(self):
383 if self._exception:
--> 384 raise self._exception
385 else:
386 return self._result
D:\Compiler\Anaconda\envs\py37\lib\site-packages\mars\deploy\oscar\session.py in init(cls, address, session_id, backend, new, timeout, **kwargs)
846 return (
847 await new_cluster_in_isolation(
--> 848 address, timeout=timeout, backend=backend, **kwargs
849 )
850 ).session
D:\Compiler\Anaconda\envs\py37\lib\site-packages\mars\deploy\oscar\local.py in new_cluster_in_isolation(address, n_worker, n_cpu, mem_bytes, cuda_devices, subprocess_start_method, backend, config, web, timeout, n_supervisor_process)
89 n_supervisor_process,
90 )
---> 91 await cluster.start()
92 return await LocalClient.create(cluster, timeout)
93
D:\Compiler\Anaconda\envs\py37\lib\site-packages\mars\deploy\oscar\local.py in start(self)
217 await self._start_worker_pools()
218 # start service
--> 219 await self._start_service()
220
221 if self._web:
D:\Compiler\Anaconda\envs\py37\lib\site-packages\mars\deploy\oscar\local.py in _start_service(self)
263 async def _start_service(self):
264 self._web = await start_supervisor(
--> 265 self.supervisor_address, config=self._config, web=self._web
266 )
267 for worker_pool, band_to_resource in zip(
D:\Compiler\Anaconda\envs\py37\lib\site-packages\mars\deploy\oscar\service.py in start_supervisor(address, lookup_address, modules, config, web)
40 config["modules"] = modules
41 try:
---> 42 await start_services(NodeRole.SUPERVISOR, config, address=address)
43 logger.debug("Mars supervisor started at %s", address)
44 except ImportError:
D:\Compiler\Anaconda\envs\py37\lib\site-packages\mars\services\core.py in start_services(node_role, config, address, mark_ready)
172 for entries in svc_entries_list:
173 instances = [svc_entry.get_instance(address, config) for svc_entry in entries]
--> 174 await asyncio.gather(*[inst.start() for inst in instances])
175
176 if mark_ready and "cluster" in service_names:
D:\Compiler\Anaconda\envs\py37\lib\site-packages\mars\services\cluster\supervisor\service.py in start(self)
65 interval=svc_config.get("node_check_interval"),
66 uid=NodeInfoUploaderActor.default_uid(),
---> 67 address=address,
68 )
69 await mo.create_actor(
D:\Compiler\Anaconda\envs\py37\lib\site-packages\mars\oscar\api.py in create_actor(actor_cls, uid, address, *args, **kwargs)
25 async def create_actor(actor_cls, *args, uid=None, address=None, **kwargs) -> ActorRef:
26 ctx = get_context()
---> 27 return await ctx.create_actor(actor_cls, *args, uid=uid, address=address, **kwargs)
28
29
D:\Compiler\Anaconda\envs\py37\lib\site-packages\mars\oscar\backends\context.py in create_actor(self, actor_cls, uid, address, *args, **kwargs)
110 future = await self._call(address, create_actor_message, wait=False)
111 result = await self._wait(future, address, create_actor_message)
--> 112 return self._process_result_message(result)
113
114 async def has_actor(self, actor_ref: ActorRef) -> bool:
D:\Compiler\Anaconda\envs\py37\lib\site-packages\mars\oscar\backends\context.py in _process_result_message(message)
74 return message.result
75 else:
---> 76 raise message.as_instanceof_cause()
77
78 async def _wait(self, future: asyncio.Future, address: str, message: _MessageBase):
D:\Compiler\Anaconda\envs\py37\lib\site-packages\mars\oscar\backends\pool.py in create_actor(self, message)
523 actor.address = address = self.external_address
524 self._actors[actor_id] = actor
--> 525 await self._run_coro(message.message_id, actor.__post_create__())
526
527 result = ActorRef(address, actor_id)
D:\Compiler\Anaconda\envs\py37\lib\site-packages\mars\oscar\backends\pool.py in _run_coro(self, message_id, coro)
341 self._process_messages[message_id] = asyncio.tasks.current_task()
342 try:
--> 343 return await coro
344 finally:
345 self._process_messages.pop(message_id, None)
D:\Compiler\Anaconda\envs\py37\lib\site-packages\mars\services\cluster\uploader.py in __post_create__(self)
58 async def __post_create__(self):
59 self._upload_task = asyncio.create_task(self._periodical_upload_node_info())
---> 60 await self._uploaded_future
61
62 async def __pre_destroy__(self):
D:\Compiler\Anaconda\envs\py37\lib\site-packages\mars\services\cluster\uploader.py in _periodical_upload_node_info(self)
84 while True:
85 try:
---> 86 await self.upload_node_info()
87 if not self._uploaded_future.done():
88 self._uploaded_future.set_result(None)
D:\Compiler\Anaconda\envs\py37\lib\site-packages\mars\services\cluster\uploader.py in upload_node_info(self, status)
120 try:
121 if not self._info.env:
--> 122 self._info.env = await asyncio.to_thread(gather_node_env)
123 self._info.detail.update(
124 await asyncio.to_thread(
D:\Compiler\Anaconda\envs\py37\lib\site-packages\mars\lib\aio\_threads.py in to_thread(func, *args, **kwargs)
34 ctx = contextvars.copy_context()
35 func_call = functools.partial(ctx.run, func, *args, **kwargs)
---> 36 return await loop.run_in_executor(None, func_call)
D:\Compiler\Anaconda\envs\py37\lib\concurrent\futures\thread.py in run(self)
55
56 try:
---> 57 result = self.fn(*self.args, **self.kwargs)
58 except BaseException as exc:
59 self.future.set_exception(exc)
D:\Compiler\Anaconda\envs\py37\lib\site-packages\mars\services\cluster\gather.py in gather_node_env()
73
74 try:
---> 75 cuda_info = mars_resource.cuda_info()
76 except NVError: # pragma: no cover
77 logger.exception("NVError encountered, cannot gather CUDA devices.")
D:\Compiler\Anaconda\envs\py37\lib\site-packages\mars\resource.py in cuda_info()
358 driver_version=driver_info.driver_version,
359 cuda_version=driver_info.cuda_version,
--> 360 products=[nvutils.get_device_info(idx).name for idx in range(gpu_count)],
361 gpu_count=gpu_count,
362 )
D:\Compiler\Anaconda\envs\py37\lib\site-packages\mars\resource.py in <listcomp>(.0)
358 driver_version=driver_info.driver_version,
359 cuda_version=driver_info.cuda_version,
--> 360 products=[nvutils.get_device_info(idx).name for idx in range(gpu_count)],
361 gpu_count=gpu_count,
362 )
D:\Compiler\Anaconda\envs\py37\lib\site-packages\mars\lib\nvutils.py in get_device_info(dev_index)
341 info = _device_infos[dev_index] = _cu_device_info(
342 index=real_dev_index,
--> 343 uuid=uuid.UUID(bytes=uuid_t.bytes),
344 name=name_buf.value.decode(),
345 multiprocessors=cores.value,
D:\Compiler\Anaconda\envs\py37\lib\uuid.py in __init__(self, hex, bytes, bytes_le, fields, int, version, is_safe)
167 if bytes is not None:
168 if len(bytes) != 16:
--> 169 raise ValueError('bytes is not a 16-char string')
170 assert isinstance(bytes, bytes_), repr(bytes)
171 int = int_.from_bytes(bytes, byteorder='big')
ValueError: [address=127.0.0.1:8482, pid=39180] bytes is not a 16-char string`
mars.new_session()
```
**Expected behavior**
session be created successfully
|
closed
|
2022-06-29T13:04:47Z
|
2023-03-31T05:00:52Z
|
https://github.com/mars-project/mars/issues/3176
|
[] |
Ukon233
| 2
|
jwkvam/bowtie
|
plotly
| 42
|
ant looks nice
|
https://ant.design/docs/react/introduce
strongly considering just always using ant.design styling for bowtie because
1. They have all the widgets I need as far as I can tell.
2. I like the UX of their components more than palantir's blueprint.
3. It looks nice.
4. More professional looking to have components with consistent look and feel.
5. Can change primary color to whatever you want.
|
closed
|
2016-11-28T21:27:34Z
|
2016-12-05T08:34:17Z
|
https://github.com/jwkvam/bowtie/issues/42
|
[] |
jwkvam
| 1
|
autogluon/autogluon
|
data-science
| 4,129
|
Make `timm` an optional dependency
|
We should see if we can make `timm` an optional dependency in the multimodal module.
- [ ] Lazy import timm only when it is used
- [ ] Make timm an optional dependency
- [ ] Ensure presets work appropriately when timm is not present
- [ ] Ensure informative error message if timm is required but not installed for a given model
|
open
|
2024-04-23T20:26:23Z
|
2024-11-02T02:14:19Z
|
https://github.com/autogluon/autogluon/issues/4129
|
[
"module: multimodal",
"dependency",
"priority: 0"
] |
Innixma
| 1
|
python-arq/arq
|
asyncio
| 417
|
Function level keep_result_s not respected for errors
|
In `finish_failed_job`, there's only a check on self.keep_result_s, not the value configured from the function. As a result, any failed tasks are stored for potentially a very long time and cannot be re-enqueued with the same job id, even if configured on the function level to not store a result.
This also relates to https://github.com/samuelcolvin/arq/issues/416 as currently it's impossible to follow this pattern for tasks that can fail, e.g. trying to re-enqueue a task that you want to have eventually succeed does nothing of the worker is configured to keep values.
|
open
|
2023-10-22T08:55:06Z
|
2023-10-22T08:55:06Z
|
https://github.com/python-arq/arq/issues/417
|
[] |
SoftMemes
| 0
|
deepset-ai/haystack
|
machine-learning
| 8,067
|
docs: clean up docstrings of InMemoryEmbeddingRetriever
|
closed
|
2024-07-24T11:01:57Z
|
2024-07-25T11:24:01Z
|
https://github.com/deepset-ai/haystack/issues/8067
|
[] |
agnieszka-m
| 0
|
|
bmoscon/cryptofeed
|
asyncio
| 740
|
There is a simple error in this line, need to put Decimal() into the left side of the / .
|
https://github.com/bmoscon/cryptofeed/blob/6bb55d30a46f50ff9d28e6297e1c0f97f4040825/cryptofeed/exchanges/binance.py#L498
|
closed
|
2021-12-10T14:21:38Z
|
2021-12-11T15:01:17Z
|
https://github.com/bmoscon/cryptofeed/issues/740
|
[] |
badosanjos
| 2
|
microsoft/nni
|
tensorflow
| 5,744
|
Not training enough times
|
Stopped training after only running 6 times, although it displayed as RUING, there was no issue with memory usage.
Ubuntu 22.04.3 LTS
python 3.8.18
nni 3.0b2





[nnimanager.log](https://github.com/microsoft/nni/files/14275177/nnimanager.log)

|
open
|
2024-02-14T04:18:00Z
|
2024-02-15T09:36:40Z
|
https://github.com/microsoft/nni/issues/5744
|
[] |
Fly-Pluche
| 1
|
Guovin/iptv-api
|
api
| 689
|
关于Docker配置问题?
|
### Don't skip these steps
- [X] I understand that I will be **blocked** if I *intentionally* remove or skip any mandatory\* field
- [X] I have checked through the search that there are no similar issues that already exist
- [X] I will not submit any issues that are not related to this project
### Occurrence environment
- [ ] Workflow
- [ ] GUI
- [X] Docker
- [ ] Command line
### Question description
我在群晖上用docker部署了项目,配置里设了 min_resolution = 1920x1080,为什么还有很多720X576的片源,另外,里面没有最小速度配置项是怎么回事。
### Related log
_No response_
|
closed
|
2024-12-15T21:43:50Z
|
2024-12-16T02:02:44Z
|
https://github.com/Guovin/iptv-api/issues/689
|
[
"question"
] |
fanghg2018
| 2
|
autokey/autokey
|
automation
| 585
|
When you clone something, the original is still highlighted instead of the clone
|
## Classification:
UI/Usability
## Reproducibility:
Always
## Version
AutoKey version: 0.95.10
Used GUI (Gtk, Qt, or both): GTK
Installed via: Mint's Software Manager
Linux Distribution: Mint Cinnamon
## Summary
Summary of the problem.
## Steps to Reproduce (if applicable)
1. Right-click on a phrase or script in the left sidebar
2. Click "Clone Item"
## Expected Results
The clone should be instantly highlighted to start doing stuff to
## Actual Results
The original remains highlighted
## Notes
Sorry I suck at programming or else I'd try to help!
|
closed
|
2021-07-23T18:27:11Z
|
2024-11-10T05:03:10Z
|
https://github.com/autokey/autokey/issues/585
|
[
"enhancement",
"autokey-gtk",
"help-wanted",
"user interface",
"easy fix"
] |
KeronCyst
| 5
|
pyg-team/pytorch_geometric
|
deep-learning
| 9,346
|
Examples folder refactor
|
### 🛠 Proposed Refactor
Hi, I am a beginner of PyG, and I get myself familiar with PyG through the documentation, now I want to try to run some examples, I felt a little confused on the current example folder in the repo. The following are my current thoughts, I put it here for your reference, looking forward to hear your feedback.
1. Is it possible to refactor the [examples folder](https://github.com/pyg-team/pytorch_geometric/tree/master/examples), now there are too many separate python scripts in the first level of the example folder, which is not helpful for beginner to get familiar with it, could you please help to tidy it?
2. There are some official tutorial examples in [documentation website](https://pytorch-geometric.readthedocs.io/en/latest/get_started/colabs.html), it looks quite helpful for beginners, could you also create a notebook folder, and put these notebooks into it?

### Suggest a potential alternative/fix
1. Classify the python scripts into several folders according to model or datasets, and provide a better readme file under example folder.
2. Create a notebook folder, and put the colab notebooks code into it.
|
open
|
2024-05-22T07:54:23Z
|
2024-05-27T07:24:22Z
|
https://github.com/pyg-team/pytorch_geometric/issues/9346
|
[
"help wanted",
"0 - Priority P0",
"refactor",
"example"
] |
zhouyu5
| 1
|
tensorpack/tensorpack
|
tensorflow
| 1,543
|
Applying quantization to the CNN for CIFAR net with BNReLU gives 0.1 validation accuracy
|
### 1. What you did:
(1) Trained the CNN in the /examples/basics for CIFAR net and added quantization layers
### 2. What you observed:
(1) The validation accuracy does not increase above 0.1

Hi,
I do not know if this should be asked here but I cannot find references to this anywhere. I trained the /examples/basics/cifar-covnet.py in DoReFaNet and the validation accuracy does not increase above 0.1.
Looking more closely, it seems that the issue is due to the batch normalization layer defined in the convnet.
Is there any specific way the batch normalization layer should be handled during quantization?

|
closed
|
2021-09-24T18:43:03Z
|
2021-09-25T14:02:34Z
|
https://github.com/tensorpack/tensorpack/issues/1543
|
[] |
Abhishek2271
| 1
|
pytest-dev/pytest-qt
|
pytest
| 211
|
blocker.all_signals_and_args is empty when using PySide2
|
First and foremost, pytest-qt has been tremendously useful. Thank you!
I am working on a UI that needs to be compatible with both PyQt5 and PySide2. When testing I noticed that with PyQt5, the all_signals_and_args seems to be populated correctly with the args from the multile signals, however, when testing with PySide2, the list is empty.
```
def test_status_complete(qtbot):
app = Application()
signals = [app.worker.status, app.worker.finished]
with qtbot.waitSignals(signals, raising=True) as blocker:
app.worker.start()
print(blocker.all_signals_and_args)
assert False
```
testing with PyQt5
```
>>> [<pytestqt.wait_signal.SignalAndArgs instance at 0x7f93c771fdd0>, <pytestqt.wait_signal.SignalAndArgs instance at 0x7f93c771fef0>]
```
testing with PySide2
```
>>> []
```
Just wanted to pass this along. Thanks again!
|
closed
|
2018-05-15T22:45:46Z
|
2020-05-14T19:43:42Z
|
https://github.com/pytest-dev/pytest-qt/issues/211
|
[
"bug :bug:"
] |
zephmann
| 4
|
huggingface/transformers
|
machine-learning
| 35,957
|
Cannot import 'GenerationOutput' in 4.48.1
|
### System Info
- `transformers` version: 4.48.1
- Platform: Linux-5.15.167.4-microsoft-standard-WSL2-x86_64-with-glibc2.31
- Python version: 3.9.5
- Huggingface_hub version: 0.28.0
- Safetensors version: 0.5.2
- Accelerate version: not installed
- Accelerate config: not found
- PyTorch version (GPU?): 2.5.1+cu124 (True)
- Tensorflow version (GPU?): not installed (NA)
- Flax version (CPU?/GPU?/TPU?): not installed (NA)
- Jax version: not installed
- JaxLib version: not installed
- Using distributed or parallel set-up in script?: <fill in>
- Using GPU in script?: <fill in>
- GPU type: NVIDIA GeForce MX450
### Who can help?
@gante
### Information
- [ ] The official example scripts
- [x] My own modified scripts
### Tasks
- [ ] An officially supported task in the `examples` folder (such as GLUE/SQuAD, ...)
- [x] My own task or dataset (give details below)
### Reproduction
```py
from transformers import AutoModelForCausalLM, AutoTokenizer, GenerationOutput
import torch
# Load model and tokenizer
model_name = "gpt2"
model = AutoModelForCausalLM.from_pretrained(model_name)
tokenizer = AutoTokenizer.from_pretrained(model_name)
# Encode input text
input_text = "Hello, how are you?"
input_ids = tokenizer(input_text, return_tensors="pt").input_ids
# Generate text (Return logits)
output = model.generate(
input_ids,
return_dict_in_generate=True,
output_scores=True,
return_logits=True
)
# Check if the output type is GenerationOutput
print(isinstance(output, GenerationOutput)) # True
```
### Expected behavior
The code above should run without any errors.
|
closed
|
2025-01-29T13:22:00Z
|
2025-03-13T08:03:51Z
|
https://github.com/huggingface/transformers/issues/35957
|
[
"bug"
] |
inthree3
| 4
|
flasgger/flasgger
|
rest-api
| 624
|
security-parameter missing for Authorization header
|
So maybe I'm doing something wrong, but I couldn't get the the Swagger-UI to actually send the Authorization header.
My configuration includes both necessary definitions imo:
```
"securityDefinitions": {
"Bearer": {
"type": "apiKey",
"name": "Authorization",
"in": "header",
"description": "Azure AD token, format 'Bearer xxyyzz'",
},
},
"security": [{"Bearer": []}],
```
I've noticed in ./flasgger/base.py that only securityDefinitions is written to self.config
```
if self.config.get("securityDefinitions"):
data["securityDefinitions"] = self.config.get(
'securityDefinitions'
)
```
on a whim I decided to just add 'security' as well - and voilà I can now send Authorization headers just fine:
```
if self.config.get("security"):
data["security"] = self.config.get("security")
```
line 421 in base.py - I've forked the project to fix it for me:
https://github.com/wiseboar/flasgger/blob/master/flasgger/base.py
|
open
|
2024-09-23T13:06:16Z
|
2024-09-23T13:09:18Z
|
https://github.com/flasgger/flasgger/issues/624
|
[] |
wiseboar
| 0
|
coqui-ai/TTS
|
pytorch
| 2,927
|
[Bug] Unable to run tts-server or server/server.py with Bark
|
### Describe the bug
Hi,
Thank you for making such excellent software.
However, maybe due to my lack of familiarity with Docker and Python, I am running into the following errors. I ran:
`docker run --rm -it -p 5002:5002 --gpus all --entrypoint /bin/bash ghcr.io/coqui-ai/tts`
`python3 TTS/server/server.py --model_name tts_models/multilingual/multi-dataset/bark --use_cuda true`
I observed an error when doing this, similar to the first large error text below. After I ran the above commands, I noticed it downloaded tens of gigabytes of files, plus used the `--rm`, which made me think it would delete the image and try to redownload them when I reran it. (Maybe it's not so good an idea to suggest to include the `--rm` in this case? I don't know, I don't know docker very well. Maybe it caches them?). So I searched how to save a container that was run with --rm. I followed the instructions and quit the terminal with `Ctrl-P Ctrl-Q`, and then ran `docker commit <CONTAINERID> tts-bark.`
When I start the container again:
`andrewdo@ai2:~$ docker run -i --gpus all -p 5002:5002 --entrypoint='/bin/bash' -t 50a9973597d3`
and when trying to run the server, I get the results below. I'm pretty sure that it's not related to my commiting to an image, since I observed the first error when initially running that command before commiting.
```
root@98e9ad432209:~# python3 TTS/server/server.py --model_name tts_models/multilingual/multi-dataset/bark --use_cuda true
> tts_models/multilingual/multi-dataset/bark is already downloaded.
Traceback (most recent call last):
File "/root/TTS/server/server.py", line 104, in <module>
synthesizer = Synthesizer(
File "/root/TTS/utils/synthesizer.py", line 93, in __init__
self._load_tts(tts_checkpoint, tts_config_path, use_cuda)
File "/root/TTS/utils/synthesizer.py", line 183, in _load_tts
self.tts_config = load_config(tts_config_path)
File "/root/TTS/config/__init__.py", line 79, in load_config
ext = os.path.splitext(config_path)[1]
File "/usr/lib/python3.10/posixpath.py", line 118, in splitext
p = os.fspath(p)
TypeError: expected str, bytes or os.PathLike object, not NoneType
root@98e9ad432209:~#
```
```
root@98e9ad432209:~# tts-server --model_name "tts_models/multilingual/multi-dataset/bark" --use_cuda true
> tts_models/multilingual/multi-dataset/bark is already downloaded.
Traceback (most recent call last):
File "/usr/local/bin/tts-server", line 33, in <module>
sys.exit(load_entry_point('TTS', 'console_scripts', 'tts-server')())
File "/usr/local/bin/tts-server", line 25, in importlib_load_entry_point
return next(matches).load()
File "/usr/lib/python3.10/importlib/metadata/__init__.py", line 171, in load
module = import_module(match.group('module'))
File "/usr/lib/python3.10/importlib/__init__.py", line 126, in import_module
return _bootstrap._gcd_import(name[level:], package, level)
File "<frozen importlib._bootstrap>", line 1050, in _gcd_import
File "<frozen importlib._bootstrap>", line 1027, in _find_and_load
File "<frozen importlib._bootstrap>", line 1006, in _find_and_load_unlocked
File "<frozen importlib._bootstrap>", line 688, in _load_unlocked
File "<frozen importlib._bootstrap_external>", line 883, in exec_module
File "<frozen importlib._bootstrap>", line 241, in _call_with_frames_removed
File "/root/TTS/server/server.py", line 104, in <module>
synthesizer = Synthesizer(
File "/root/TTS/utils/synthesizer.py", line 93, in __init__
self._load_tts(tts_checkpoint, tts_config_path, use_cuda)
File "/root/TTS/utils/synthesizer.py", line 183, in _load_tts
self.tts_config = load_config(tts_config_path)
File "/root/TTS/config/__init__.py", line 79, in load_config
ext = os.path.splitext(config_path)[1]
File "/usr/lib/python3.10/posixpath.py", line 118, in splitext
p = os.fspath(p)
TypeError: expected str, bytes or os.PathLike object, not NoneType
```
Any idea how to get the TTS (w/Bark) server working? I haven't changed the config as far as I remember.
I did note that this did work:
`tts --text "This is a test" --model_name "tts_models/multilingual/multi-dataset/bark" --use_cuda true --out_path "/tmp/example.wav"`
However, I'm trying to get Bark to work with Rhasspy as a Local Command (or Remote HTTP) TTS server, so I need some kind of server functionality.
Thanks,
Andrew
### To Reproduce
docker run --rm -it -p 5002:5002 --gpus all --entrypoint /bin/bash ghcr.io/coqui-ai/tts
python3 TTS/server/server.py --model_name tts_models/multilingual/multi-dataset/bark --use_cuda true
### Expected behavior
_No response_
### Logs
_No response_
### Environment
```shell
python3 ./TTS/bin/collect_env_info.py
{
"CUDA": {
"GPU": [
"NVIDIA GeForce RTX 3090"
],
"available": true,
"version": "11.8"
},
"Packages": {
"PyTorch_debug": false,
"PyTorch_version": "2.0.1+cu118",
"TTS": "0.16.6",
"numpy": "1.22.0"
},
"System": {
"OS": "Linux",
"architecture": [
"64bit",
""
],
"processor": "x86_64",
"python": "3.10.12",
"version": "#1 SMP Debian 5.10.140-1 (2022-09-02)"
}
}
```
### Additional context
_No response_
|
closed
|
2023-09-05T16:52:47Z
|
2025-01-15T16:57:36Z
|
https://github.com/coqui-ai/TTS/issues/2927
|
[
"bug",
"help wanted",
"wontfix"
] |
aindilis
| 9
|
SALib/SALib
|
numpy
| 207
|
Unable to install SALib
|
When I execute 'pip install SALib' on centOS 7, it is giving an error of "distutils.errors.DistutilsError: Could not find suitable distribution for Requirement.parse('pyscaffold<2.6a0,>=2.5a0')".
After installed pyscaffold (2.5.2), it is giving error of "your setuptools version is too old(<12)",but my setuptools version is 39.1.0,and this is the biggest version required by tensorflow which is my necessary.
However , everything go well on windows.
|
closed
|
2018-09-11T08:08:52Z
|
2019-03-14T15:45:55Z
|
https://github.com/SALib/SALib/issues/207
|
[] |
valwu
| 6
|
dmlc/gluon-nlp
|
numpy
| 1,242
|
[Numpy Refactor] Tokenizers wishlist
|
We revised the implementation of tokenizers in the new version of GluonNLP.
Basically we have integrated the following tokenizers:
- whitespace
- spacy
- jieba
- SentencePiece
- YTTM
- HuggingFaceBPE
- HuggingFaceByteBPE
- HuggingFaceWordPiece
For all tokenizers, we support the following methods:
- Encode into a list of integers: `encode('hello world!', int)`
- Encode into a list of string tokens: `encode('hello world!', str)`
- Encode into a list of integers + offsets: `encode_with_offsets('hello world!', int)`
- Encode into a list of string tokens + offsets: `encode_with_offsets('hello world!', str)`
To give an example, we load the tokenizer in ALBERT, which is a SentencePieceTokenizer and illustrate these functionalities:
```python
In [1]: from gluonnlp.models.albert import get_pretrained_albert
In [2]: cfg, tokenizer, _,_ = get_pretrained_albert()
In [3]: tokenizer
Out[3]:
SentencepieceTokenizer(
model_path = /Users/xjshi/.mxnet/models/nlp/google_albert_base_v2/spm-65999e5d.model
do_lower = True, nbest = 0, alpha = 0.0
vocab = Vocab(size=30000, unk_token="<unk>", pad_token="<pad>", cls_token="[CLS]", sep_token="[SEP]", mask_token="[MASK]")
)
In [4]: tokenizer.encode('hello world!', int)
Out[4]: [10975, 126, 187]
In [5]: tokenizer.encode('hello world!', str)
Out[5]: ['▁hello', '▁world', '!']
In [6]: tokenizer.encode_with_offsets('hello world!', str)
Out[6]: (['▁hello', '▁world', '!'], [(0, 5), (5, 11), (11, 12)])
In [7]: tokenizer.encode_with_offsets('hello world!', int)
Out[7]: ([10975, 126, 187], [(0, 5), (5, 11), (11, 12)])
```
However, there are a lot of other commonly used tokenizers. We can consider to integrate:
- [ ] BlingFire: https://github.com/microsoft/BlingFire
- [ ] Mecab: A commonly used tokenizer for Japanese: https://taku910.github.io/mecab/
|
open
|
2020-06-10T07:25:57Z
|
2020-06-10T16:13:11Z
|
https://github.com/dmlc/gluon-nlp/issues/1242
|
[
"enhancement",
"numpyrefactor"
] |
sxjscience
| 0
|
microsoft/nni
|
data-science
| 4,783
|
please add the ppo_tuner performance in comparison of hpo algorithms
|
**Describe the issue**:
Hi,
Could you please add the ppo_tuner performance in comparison of hpo algorithms:
https://nni.readthedocs.io/en/latest/sharings/hpo_comparison.html
Thanks a lot!
**Environment**:
- NNI version:
- Training service (local|remote|pai|aml|etc):
- Client OS:
- Server OS (for remote mode only):
- Python version:
- PyTorch/TensorFlow version:
- Is conda/virtualenv/venv used?:
- Is running in Docker?:
**Configuration**:
- Experiment config (remember to remove secrets!):
- Search space:
**Log message**:
- nnimanager.log:
- dispatcher.log:
- nnictl stdout and stderr:
<!--
Where can you find the log files:
LOG: https://github.com/microsoft/nni/blob/master/docs/en_US/Tutorial/HowToDebug.md#experiment-root-director
STDOUT/STDERR: https://github.com/microsoft/nni/blob/master/docs/en_US/Tutorial/Nnictl.md#nnictl%20log%20stdout
-->
**How to reproduce it?**:
|
open
|
2022-04-20T13:18:47Z
|
2022-04-24T08:56:03Z
|
https://github.com/microsoft/nni/issues/4783
|
[] |
southisland
| 1
|
FlareSolverr/FlareSolverr
|
api
| 297
|
Can't access property "documentTitle", this.browsingContext.currentWindowGlobal is null
|
This issue is reported with FlareSolverr 2.2.0 in Windows and CentOS.
I think it's not related to user permissions but a Firefox bug. We are using the latest nightly.
I'm not able to reproduce the issue, just once.
Related #295 #286 #289 #282
|
closed
|
2022-02-06T10:56:15Z
|
2023-01-05T11:19:13Z
|
https://github.com/FlareSolverr/FlareSolverr/issues/297
|
[
"more information needed",
"confirmed"
] |
ngosang
| 3
|
miguelgrinberg/Flask-Migrate
|
flask
| 456
|
Add timezone support
|
#### What's the problem this feature will solve?
If you try to generate a migration that requires a DateTime with timezone support you'll get the following error:
```
ERROR: MainProcess<flask_migrate> Error: The library 'python-dateutil' is required for timezone support
```
It seems `python-dateutil` is optional dependency since [alembic==1.7.0](https://alembic.sqlalchemy.org/en/latest/changelog.html#change-c01e812b3ac23a262b2c9c343f009aad) and can be installed via extras:`pip install alembic[tz]`.
#### Describe the solution you'd like
Would it make sense to reflect this change to `flask-migrate` by adding `flask-migrate[tz]` extras which would require `alembic[tz]`?
#### Alternative Solutions
Add manually `alembic[tz]` or even `python-dateutil` to pip-tools/poetry/pipenv/reqirements.txt requirement flies.
#### Additional context
- https://github.com/sqlalchemy/alembic/issues/674#issuecomment-604585944
- https://github.com/sqlalchemy/alembic/blob/a9e6f9079a7d0157a842b3786ebd4f7031b08912/setup.cfg#L53-L55
P.S. I'd like to shoot a PR if the maintainer is on board.
|
closed
|
2022-03-16T12:07:10Z
|
2022-03-16T15:21:40Z
|
https://github.com/miguelgrinberg/Flask-Migrate/issues/456
|
[
"question"
] |
atugushev
| 2
|
deezer/spleeter
|
tensorflow
| 493
|
[Discussion] updating Python dependencies
|
Some Python packages listed in `setup.py` are a little bit old:
- pandas is now `1.1.2` as opposed to `0.25.1`
- librosa is now `0.8.0` as opposed to `0.7.2`
- it now requires `numba==0.48.0` for itself, so unless you're using numba in the actual project, you can remove it
Also, I think you can:
- safely replace all current `==` conditions to `>=` for all items in `install_requires`
- maybe add a condition like `tensorflow >=1.14, <2.0` so that it will work with tensorflow 1.14 and avoid [issues like this](https://stackoverflow.com/questions/61491893/i-cannot-install-tensorflow-version-1-15-through-pip)
- maybe add support for python 3.8 or even the [upcoming 3.9](https://docs.python.org/3.9/whatsnew/3.9.html)
|
closed
|
2020-09-13T20:23:43Z
|
2020-10-09T15:32:56Z
|
https://github.com/deezer/spleeter/issues/493
|
[
"question"
] |
flriancu
| 3
|
flaskbb/flaskbb
|
flask
| 170
|
Performance issues
|
I am creating a general topic for this and posting the first performance issue I encountered.
Using standard install in production mode with redis configured I am seeing very consistently slow response times. With plain "hello world" nginx + uwsgi the response time is 0.309 ms
When using flask BB the response time is average 3 seconds. I am testing over LAN using raspbian minimal and narrowed it down to flaskbb alone taking that amount of time in processing a request for the index page.
What information do you guys need? I can code some flask (not much) so I am here to help.
I will try a development install and use Werkzeug profiling to see where the slowness is.
|
closed
|
2016-01-09T14:35:01Z
|
2018-04-15T07:47:37Z
|
https://github.com/flaskbb/flaskbb/issues/170
|
[] |
abshkd
| 6
|
chaoss/augur
|
data-visualization
| 2,752
|
Stability of our production SAAS instance of Augur
|
As we look to expand our SAAS offerings within CHAOSS, I wonder if we could get some additional funding that would allow us to run our "production" instance in a completely separate environment that wouldn't be impacted by issues like the one we just had with a dev instance getting overloaded and taking other instances down?
The other thing that would be helpful once we have a production environment is advance notification of ALL changes that are made on that instance or maybe maintenance windows when we can expect changes that might impact usage?
|
open
|
2024-03-26T16:30:34Z
|
2024-03-28T16:54:09Z
|
https://github.com/chaoss/augur/issues/2752
|
[
"feature-request",
"deployed version",
"CHAOSS",
"documentation",
"usability",
"server",
"database",
"add-feature",
"devops"
] |
geekygirldawn
| 1
|
thp/urlwatch
|
automation
| 97
|
how to enable color diffs?
|
Hi,
What is the configuration to enable color diffs? I don't find it in the readme.
Thanks.
|
closed
|
2016-09-30T11:01:28Z
|
2016-10-02T09:58:46Z
|
https://github.com/thp/urlwatch/issues/97
|
[] |
monperrus
| 1
|
praw-dev/praw
|
api
| 2,004
|
Installing praw from source or sdist fails due to importing runtime deps at build time
|
### Describe the Bug
Same issue as praw-dev/prawcore#164 and @LilSpazJoekp requested followup from praw-dev/prawcore#165
Due to the use of Flit's fallback automatic metadata version extraction needing to dynamically import the package `__init__.py` instead of reading it statically (see pypa/flit#386 ) , since the actual `__version__` is imported from `const.py`, this requires the runtime dependencies be installed in the build environment when building or installing from an sdist (or the source tree).
This happens merely by chance to currently be a transitive backend dependency of `flit_core` and thus building in isolated mode works. However, this shouldn't be relied upon, as if either `flit_core` or `praw`'s dependencies were to ever change, this would break isolated builds too. And `requests` _isn't_ an exposed non-backend dependency of `flit-core`, so it doesn't actually get installed otherwise.
This means Requests must be manually installed (not just the explicitly pyproject,toml-specified build dependencies) if building/installing `praw` in any other context than `pip`'s isolated mode, e.g. without the `--no-build-isolation` flag, via the legacy non-PEP 517 builder, or via other tools or most downstream ecosystems that use other build isolation mechanisms. In particular, this was an issue on conda-forge/prawcore-feedstock#14 where I was packaging the new `prawcore` 2.4.0 version for Conda-Forge—you can [see the full Azure build log](https://dev.azure.com/conda-forge/feedstock-builds/_build/results?buildId=806405&view=logs&jobId=656edd35-690f-5c53-9ba3-09c10d0bea97&j=656edd35-690f-5c53-9ba3-09c10d0bea97&t=986b1512-c876-5f92-0d81-ba851554a0a3).
This can be worked around for now by manually installing `requests` into the build environment, but that's certainly not an ideal solution as it adds an extra build dependency (and its dependencies in turn), requires extra work by everyone installing from source (without build isolation), makes builds take longer, and is fragile and not easily maintainable/scalable long term for other runtime dependencies.
### Desired Result
Praw is able to be built and installed without installing requests, or relying on it happening to be a transitive build backend dependency of `flit_core` and present "by accident".
While there are other ways to achieve this, as described in praw-dev/prawcore#164 , @LilSpazJoekp indicated he preferred moving `__version__` to be in `__init__.py` directly, so Flit can find it by static inspection without triggering an import, and naturally asked for the same approach here as applied in praw-dev/prawcore#165 .
This works, with one additional complication: the [`USER_AGENT_FORMAT` module-level constant in `const.py`](https://github.com/praw-dev/praw/blob/ec3b6caa79d838b45cd4af18b925dcca2e68e2e6/praw/const.py#L6) relies in turn on `__version__`, which means that `praw.const` needs to import it from `praw` (`__init__`), and because `praw.reddit.Reddit` is imported in `__init__` for convenience (which in turn imports `praw.const`), this creates an import cycle (as well as making `praw` expensive to import, on the order of multiple seconds on an older machine).
This cannot be trivially solved by a scope-local import, because `USER_AGENT_FORMAT` is a module-level constant. The simplest and likely least disruptive solution, since `USER_AGENT_FORMAT` is only used on place in PRAW (`praw.reddit`) is to just inject `__version__` at the one point of use along with the user-configured user agent rather than statically, eliminating an extra layer of format replacement nesting in the process.
The one non-trivial impact from this is requiring a change to user code directly using `USER_AGENT_FORMAT` themselves to insert `praw.__version__` along with their own user agent. After [various grep.app queries](https://grep.app/search?q=praw.const&case=true&filter[lang][0]=Python), which scans the entirety of public code on GitHub, I did find [a single instance of this](https://github.com/dickmao/nnreddit/blob/7bb466aef038fd6e1d3155c7b33953dbe24fab48/nnreddit/authenticated_reddit.py#L90) that would be affected, in the (closed-development) [nnreddit](https://github.com/dickmao/nnreddit) project providing a Reddit backend for the Gnus reader. However, the fix is straightforward (an extra arg to one `format` call) and backward-compatible with older PRAW, and the primary change here, removing, `__version__` from `praw.const` is also equally technically backward-incompatible and was accepted in `prawcore`, so not sure how much of an issue that is for you.
Using a named placeholder for the PRAW version, e.g.`"{} PRAW/{praw_version}"`, would give a clearer error message as to what is needed to add to the `format` call, while modifying existing code (e.g. `USER_AGENT_FORMAT.format(user_agent)`) to support the new PRAW version (i.e. `USER_AGENT_FORMAT.format(user_agent, praw_version=praw.__version__)` would be fully backward compatible with `USER_AGENT_FORMAT` in older PRAW, as `format` will simply drop arguments that don't match placeholders in the string.
The other alternative as mentioned in the other issue would be simply setting the version statically in `pyproject.toml`. This would avoid all these issues and needing to change anything else besides [the `set_version.py` script](https://github.com/praw-dev/praw/blob/ec3b6caa79d838b45cd4af18b925dcca2e68e2e6/tools/set_version.py) as well as give you the benefits of static metadata. It would mean the version is defined multiple places which you wanted to avoid, but if you're using that script to set it anyway, it shouldn't actually require any additional maintainer effort since the script could take care of it too. Of course, if that's the case we'd presumably want to go back and make that same change in `prawcore` too; however, it would simplify the implementation in `asyncpraw` (as well as `asyncprawcore`) and avoid this same issue with `USER_AGENT_FORMAT`, and for which `asyncpraw.const.__version__` is used at least [once in the wild](https://github.com/kaif-00z/TgRedditBot/blob/987a767d486976a4fe259a80484386a4165c9ed4/bot.py#L136).
Up to you!
### Code to reproduce the bug
In a fresh venv without requests or praw installed, e.g.
```bash
python3 -m venv .venv
source .venv/bin/activate
pip install --upgrade pip wheel
```
Run:
```bash
pip install flit-core # Or flit
pip install --no-build-isolation .
```
### My code does not include sensitive credentials
- [X] Yes, I have removed sensitive credentials from my code.
### Relevant Logs
```Shell
See praw-dev/prawcore#164
```
### This code has previously worked as intended
Yes
### Operating System/Environment
Any
### Python Version
Any, tested 3.9-3.12
### PRAW Version
7.7.2.dev0
### Links, references, and/or additional comments?
_No response_
|
open
|
2023-12-15T22:19:44Z
|
2025-03-17T06:26:44Z
|
https://github.com/praw-dev/praw/issues/2004
|
[] |
CAM-Gerlach
| 12
|
pydata/xarray
|
numpy
| 9,653
|
Dataset.to_dataframe() dimension order is not alphabetically sorted by default
|
### What happened?
Hi, I noticed that the [documentation](https://docs.xarray.dev/en/stable/generated/xarray.Dataset.to_dataframe.html) for `Dataset.to_dataframe()` says that "by default, dimensions are sorted alphabetically". This is contrast with [`DataArray.to_dataframe()`](https://docs.xarray.dev/en/stable/generated/xarray.DataArray.to_dataframe.html), where the order is given by the order of the dimensions in the `DataArray`, which was discussed [in this comment](https://github.com/pydata/xarray/pull/4333#issuecomment-672084936).
However, it appears that `Dataset.to_dataframe()` doesn't in fact sort the orders alphabetically with this example on current main 8f6e45ba:
```python
import xarray as xr
ds = xr.Dataset({
"foo": xr.DataArray(0, coords=[("y", [1, 2, 3]), ("x", [4, 5, 6])]),
})
print(ds.to_dataframe())
```
I get
```
foo
y x
1 4 0
5 0
6 0
2 4 0
5 0
6 0
3 4 0
5 0
6 0
```
### What did you expect to happen?
The dimensions in the output should be sorted alphabetically, like this:
```
foo
x y
4 1 0
2 0
3 0
5 1 0
2 0
3 0
6 1 0
2 0
3 0
```
### Minimal Complete Verifiable Example
```Python
See above
```
### MVCE confirmation
- [X] Minimal example — the example is as focused as reasonably possible to demonstrate the underlying issue in xarray.
- [X] Complete example — the example is self-contained, including all data and the text of any traceback.
- [X] Verifiable example — the example copy & pastes into an IPython prompt or [Binder notebook](https://mybinder.org/v2/gh/pydata/xarray/main?urlpath=lab/tree/doc/examples/blank_template.ipynb), returning the result.
- [X] New issue — a search of GitHub Issues suggests this is not a duplicate.
- [X] Recent environment — the issue occurs with the latest version of xarray and its dependencies.
### Relevant log output
_No response_
### Anything else we need to know?
_No response_
### Environment
<details>
INSTALLED VERSIONS
------------------
commit: None
python: 3.12.7 (main, Oct 1 2024, 00:00:00) [GCC 14.2.1 20240912 (Red Hat 14.2.1-3)]
python-bits: 64
OS: Linux
OS-release: 6.11.3-200.fc40.x86_64
machine: x86_64
processor:
byteorder: little
LC_ALL: None
LANG: en_US.UTF-8
LOCALE: ('en_US', 'UTF-8')
libhdf5: None
libnetcdf: None
xarray: 2024.9.1.dev73+g8f6e45ba
pandas: 2.2.3
numpy: 1.26.4
scipy: None
netCDF4: None
pydap: None
h5netcdf: None
h5py: None
zarr: None
cftime: None
nc_time_axis: None
iris: None
bottleneck: None
dask: None
distributed: None
matplotlib: None
cartopy: None
seaborn: None
numbagg: None
fsspec: None
cupy: None
pint: None
sparse: None
flox: None
numpy_groupies: None
setuptools: 69.0.3
pip: 24.0
conda: None
pytest: None
mypy: None
IPython: None
sphinx: None
</details>
|
closed
|
2024-10-21T11:33:00Z
|
2024-10-23T08:09:16Z
|
https://github.com/pydata/xarray/issues/9653
|
[
"topic-documentation"
] |
mgunyho
| 4
|
Farama-Foundation/PettingZoo
|
api
| 600
|
StopIteration
|
Hi, i have a problem and i don't know how to solve it, could you give me some advice?
File "/home/user/anaconda3/lib/python3.6/site-packages/supersuit/utils/make_defaultdict.py", line 5, in make_defaultdict
dd = defaultdict(type(next(iter(d.values()))))
StopIteration
|
closed
|
2022-01-01T17:14:00Z
|
2022-01-28T17:38:43Z
|
https://github.com/Farama-Foundation/PettingZoo/issues/600
|
[] |
xiaominy
| 3
|
autogluon/autogluon
|
scikit-learn
| 4,314
|
Error during TabularPredictor's `predict_proba`
|
I am using `TabularPredictor` for a classification problem. I am using the method `TabularPredictor.predict`, which throws the error:
```
OSError: libgomp.so.1: cannot open shared object file: No such file or directory
```
On further reading the traceback, the problem starts when it internally calls `predict_proba`.
The problem exists even after installing the `libgomp` library on Linux.
The predict method works fine in case of regression problems.
I am using `autogluon==0.8.2`
|
closed
|
2024-07-08T05:02:51Z
|
2024-07-10T05:06:16Z
|
https://github.com/autogluon/autogluon/issues/4314
|
[
"module: tabular",
"OS: Mac",
"install"
] |
ArijitSinghEDA
| 3
|
Farama-Foundation/PettingZoo
|
api
| 434
|
Pygame rendering/observing returns a reference rather than a new object in Cooperative Pong
|
According to the pygame documentation for pygame.surfarray.pixels3d:
> Create a new 3D array that directly references the pixel values in a Surface. Any changes to the array will affect the pixels in the Surface. This is a fast operation since no data is copied.
Any environment with a render function that directly returns the result of this method returns a reference to an object that is controlled and can still be mutated by pygame.
This can cause issues, for example, if you save all of the renders to an array, you would expect the array to be populated by the individual renders at each step, but the array will actually be full of references to the latest render.
I searched through all references to that function in PettingZoo, and this issue only affects Cooperative Pong (render and observe).
|
closed
|
2021-07-30T18:20:45Z
|
2021-07-30T18:30:54Z
|
https://github.com/Farama-Foundation/PettingZoo/issues/434
|
[] |
RyanNavillus
| 0
|
voxel51/fiftyone
|
computer-vision
| 5,492
|
[FR] Support more image codecs in `fiftyone.utils.image.read`
|
### Proposal Summary
Support more codecs when reading images from `fiftyone.utils.image.read`
### What areas of FiftyOne does this feature affect?
- [ ] App: FiftyOne application
- [x] Core: Core `fiftyone` Python library
- [ ] Server: FiftyOne server
### Details
`read` uses opencv, which doesn't support `.avif` images (well at least not from what I could see):
Example 1:
```py
from fiftyone.utils.image import read
url = "https://aomediacodec.github.io/av1-avif/testFiles/Link-U/hato.profile0.8bpc.yuv420.no-cdef.avif"
img = read(url)
```
```py
Traceback (most recent call last):
File "/home/laurent/Documents/project/scratch.py", line 810, in <module>
img = read(url)
^^^^^^^^^
File "/home/laurent/Documents/project/.venv/lib/python3.12/site-packages/fiftyone/utils/image.py", line 37, in read
return etai.read(path_or_url, include_alpha=include_alpha, flag=flag)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/home/laurent/Documents/project/.venv/lib/python3.12/site-packages/eta/core/image.py", line 476, in read
return download(path_or_url, include_alpha=include_alpha, flag=flag)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/home/laurent/Documents/project/.venv/lib/python3.12/site-packages/eta/core/image.py", line 457, in download
return decode(bytes, include_alpha=include_alpha, flag=flag)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/home/laurent/Documents/project/.venv/lib/python3.12/site-packages/eta/core/image.py", line 438, in decode
return _exchange_rb(cv2.imdecode(vec, flag))
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/home/laurent/Documents/project/.venv/lib/python3.12/site-packages/eta/core/image.py", line 1982, in _exchange_rb
if is_gray(img):
^^^^^^^^^^^^
File "/home/laurent/Documents/project/.venv/lib/python3.12/site-packages/eta/core/image.py", line 1297, in is_gray
return img.ndim == 2 or (img.ndim == 3 and img.shape[2] == 1)
^^^^^^^^
AttributeError: 'NoneType' object has no attribute 'ndim'
```
Example 2:
```bash
$ file test.avif
test.avif: ISO Media, AVIF Image
```
```py
from fiftyone.utils.image import read
filepath = "test.avif"
img = read(filepath)
```
```py
Traceback (most recent call last):
File "/home/laurent/Documents/project/scratch.py", line 810, in <module>
img = read(filepath)
^^^^^^^^^^^^^^
File "/home/laurent/Documents/project/.venv/lib/python3.12/site-packages/fiftyone/utils/image.py", line 37, in read
return etai.read(path_or_url, include_alpha=include_alpha, flag=flag)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/home/laurent/Documents/project/.venv/lib/python3.12/site-packages/eta/core/image.py", line 481, in read
raise OSError("Image not found '%s'" % path_or_url)
OSError: Image not found 'test.avif'
```
The errors from the traceback aren't also very helpful.
### Workaround
In my usecase, I encounter this issue because I use `fiftyone.Dataset.apply_model` (which calls internally `fiftyone.utils.image.read`).
Adding an option to the internal of `fiftyone.Dataset.apply_model` to forward only the sample to the model could work. Essentially adding a `only_needs_sample` argument or something here:
https://github.com/voxel51/fiftyone/blob/f40ab0ee48a772008e2e79fe66a16eb31f5501e0/fiftyone/core/models.py#L328-L334
Since I'm creating my own model, I can handle the loading of the image myself.
### Willingness to contribute
The FiftyOne Community welcomes contributions! Would you or another member of your organization be willing to contribute an implementation of this feature?
- [ ] Yes. I can contribute this feature independently
- [x] Yes. I would be willing to contribute this feature with guidance from the FiftyOne community
- [ ] No. I cannot contribute this feature at this time
|
open
|
2025-02-14T14:24:48Z
|
2025-02-14T16:53:14Z
|
https://github.com/voxel51/fiftyone/issues/5492
|
[
"feature"
] |
Laurent2916
| 1
|
ymcui/Chinese-BERT-wwm
|
nlp
| 82
|
Chinese-BERT-wwm与Chinese-PreTrained-XLNet的模型下载地址有错误
|
其中两者的base版模型的讯飞云下载地址发生了交错。
|
closed
|
2020-01-01T22:29:18Z
|
2020-01-02T00:41:31Z
|
https://github.com/ymcui/Chinese-BERT-wwm/issues/82
|
[] |
fungxg
| 1
|
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.