repo_name
stringlengths 9
75
| topic
stringclasses 30
values | issue_number
int64 1
203k
| title
stringlengths 1
976
| body
stringlengths 0
254k
| state
stringclasses 2
values | created_at
stringlengths 20
20
| updated_at
stringlengths 20
20
| url
stringlengths 38
105
| labels
listlengths 0
9
| user_login
stringlengths 1
39
| comments_count
int64 0
452
|
|---|---|---|---|---|---|---|---|---|---|---|---|
Gozargah/Marzban
|
api
| 1,320
|
Errors in latest dev version
|
When I try to search a user in the panel, if there are multiple instances only 10 of them will show and I get this error in logs.

also this error appears in the logs when some user ( I do not know which ) updates his subscription.

|
closed
|
2024-09-17T12:24:24Z
|
2024-09-19T13:00:28Z
|
https://github.com/Gozargah/Marzban/issues/1320
|
[
"Question"
] |
mhmdh94
| 1
|
biolab/orange3
|
data-visualization
| 6,837
|
qt.svg: Cannot open file '<local path>/canvas_icons:/Dropdown.svg'
|
When trying to run the env with "python -m Orange.canvas", this alert arises:
qt.svg: Cannot open file '<local path>/canvas_icons:/Dropdown.svg', because: La sintassi del nome del file, della directory o del volume non ? corretta.
(in english: file name syntax, directory or volume is wrong)
<!--
Thanks for taking the time to report a bug!
If you're raising an issue about an add-on (i.e., installed via Options > Add-ons), raise an issue in the relevant add-on's issue tracker instead. See: https://github.com/biolab?q=orange3
To fix the bug, we need to be able to reproduce it. Please answer the following questions to the best of your ability.
-->
**What's wrong?**
<!-- Be specific, clear, and concise. Include screenshots if relevant. -->
<!-- If you're getting an error message, copy it, and enclose it with three backticks (```). -->
**How can we reproduce the problem?**
<!-- Upload a zip with the .ows file and data. -->
<!-- Describe the steps (open this widget, click there, then add this...) -->
**What's your environment?**
<!-- To find your Orange version, see "Help → About → Version" or `Orange.version.full_version` in code -->
- Operating system:
- Orange version:
- How you installed Orange:
|
closed
|
2024-06-17T09:18:54Z
|
2024-08-30T08:02:45Z
|
https://github.com/biolab/orange3/issues/6837
|
[
"bug"
] |
Alex72RM
| 3
|
satwikkansal/wtfpython
|
python
| 348
|
Generate Jupyter notebooks for all translations
|
After #343 is done and translations are synced with base repo, translation maintainers shall generate `Jyputer` notebook
|
open
|
2024-10-16T08:04:27Z
|
2024-10-16T08:04:27Z
|
https://github.com/satwikkansal/wtfpython/issues/348
|
[] |
nifadyev
| 0
|
huggingface/transformers
|
pytorch
| 36,290
|
past_key_value(s) name inconsistency causing problems
|
### System Info
- `transformers` version: 4.50.0.dev0
- Platform: Linux-6.4.3-0_fbk14_zion_2601_gcd42476b84e9-x86_64-with-glibc2.34
- Python version: 3.12.9
- Huggingface_hub version: 0.28.1
- Safetensors version: 0.5.2
- Accelerate version: 1.4.0
- Accelerate config: not found
- DeepSpeed version: not installed
- PyTorch version (GPU?): 2.6.0.dev20241112+cu121 (True)
- Tensorflow version (GPU?): not installed (NA)
- Flax version (CPU?/GPU?/TPU?): not installed (NA)
- Jax version: not installed
- JaxLib version: not installed
- Using distributed or parallel set-up in script?: no
- Using GPU in script?: yes
- GPU type: NVIDIA H100
### Who can help?
@ArthurZucker probably others
### Information
- [x] The official example scripts
- [ ] My own modified scripts
### Tasks
- [x] An officially supported task in the `examples` folder (such as GLUE/SQuAD, ...)
- [ ] My own task or dataset (give details below)
### Reproduction
run https://huggingface.co/docs/transformers/main/en/quantization/torchao
### Expected behavior
no error
____
this error is related to https://github.com/huggingface/transformers/pull/36289
a bunch of models use past_key_value and past_key_values interchangeably, this causes issues since kwarg names are hardcoded to be skipped by the _skip_keys_device_placement attribute. This causes issues any time torch.compile is used with a model that has this issue.
The above PR fixes the issue for llama but other models like src/transformers/models/moonshine/modeling_moonshine.py
src/transformers/models/mistral/modeling_mistral.py
src/transformers/models/emu3/modeling_emu3.py
...etc, have the same issue which is actually breaking CI for that PR.
this is also the cause of https://github.com/pytorch/ao/issues/1705 which is where this was first surfaced.
is there a reason for these two names to be used instead of just one? If not it seems like they should be entirely consolidated to avoid such issues, if so, then _skip_keys_device_placement needs to include both across all models or
|
open
|
2025-02-19T22:03:42Z
|
2025-03-22T08:03:05Z
|
https://github.com/huggingface/transformers/issues/36290
|
[
"bug"
] |
HDCharles
| 1
|
akfamily/akshare
|
data-science
| 5,941
|
AKShare 接口问题报告 - stock_board_industry_index_ths
|
已经升级到了AKShare最新版:akshare 1.16.58
Python版本:Python 3.12.8
操作系统:Mac 14.6.1
报错接口:stock_board_industry_index_ths
问题描述:
通过stock_board_concept_name_ths获得同花顺的概念板块列表,
然后通过stock_board_industry_index_ths获得板块指数,发现大部分的板块都无法正常获取
代码如下:
```
import akshare as ak
sklist = ak.stock_board_concept_name_ths()
print(sklist)
stock_board_industry_index_ths_df = ak.stock_board_industry_index_ths(symbol="自由贸易港", start_date="20250301", end_date="20250305")
print(stock_board_industry_index_ths_df)
```
console返回:
```
name code
0 阿尔茨海默概念 308614
1 AI PC 309121
2 AI手机 309120
3 AI语料 309126
4 阿里巴巴概念 301558
.. ... ...
356 租售同权 302034
357 自由贸易港 306398
358 3D打印 300127
359 5G 300843
360 6G概念 309055
[361 rows x 2 columns]
Traceback (most recent call last):
File "/1.py", line 6, in <module>
stock_board_industry_index_ths_df = ak.stock_board_industry_index_ths(symbol="阿里巴巴概念", start_date="20250301", end_date="20250305")
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/Users/lord/.pyenv/versions/3.12.8/lib/python3.12/site-packages/akshare/stock_feature/stock_board_industry_ths.py", line 139, in stock_board_industry_index_ths
symbol_code = code_map[symbol]
~~~~~~~~^^^^^^^^
KeyError: '阿里巴巴概念'
```
目前试过了:阿尔茨海默概念、AI PC、AI手机、阿里巴巴概念租售同权、自由贸易港 等都无法正确获得数据。
|
closed
|
2025-03-21T01:26:21Z
|
2025-03-21T10:07:47Z
|
https://github.com/akfamily/akshare/issues/5941
|
[
"bug"
] |
1eez
| 2
|
HumanSignal/labelImg
|
deep-learning
| 261
|
Licensing?
|
While working on a feature for https://github.com/wkentaro/labelme, I (accidentally) copied a line of code from that project into a Google Search. It seems that there's quite some overlap between the code of wkentaro's labelme project and the code in this project.
After some more reading, I found that wkentaro's labelme project put [an acknowledgement](https://github.com/wkentaro/labelme#acknowledgement) to https://github.com/mpitid/pylabelme. That project was last touched in 2011 and again has large parts of code that are exactly the same. It seems that mpitid's pylabelme project is not only the origin of wkentaro's labelme project, but also this project. Is that true?
|
open
|
2018-03-24T22:37:35Z
|
2018-03-24T22:37:35Z
|
https://github.com/HumanSignal/labelImg/issues/261
|
[] |
mbuijs
| 0
|
kaliiiiiiiiii/Selenium-Driverless
|
web-scraping
| 132
|
(selenium-driverless 1.7)await driver.find_element(By.XPATH,'xxx') : Unable to find element
|
closed
|
2023-12-16T17:57:03Z
|
2023-12-16T18:01:28Z
|
https://github.com/kaliiiiiiiiii/Selenium-Driverless/issues/132
|
[] |
User-Clb
| 1
|
|
numba/numba
|
numpy
| 9,886
|
numba jit: "Type of variable <varname>.2 cannot be determined" for a fully specified variable.
|
# Bug report
### Bug description:
I have several functions that work fine with jit and njit. All variables are explicitly typed, such as
```python
def afun(parm1:int, parm2:int)->list[int]:
varname:int=3
listname:list[int]=[0]
```
In one function, the jit complains about a variable as having an undeterminable type, even though it has been explicitly typed:
```Type of variable 'artp.2' cannot be determined, operation: call $1078load_global.2($binop_add1104.10, func=$1078load_global.2, args=[Var($binop_add1104.10, primes2025a.py:240)], kws=(), vararg=None, varkwarg=None, target=None), location: /home/dakra/./primes2025a.py (240)```
where the code looks like:
```python
from numba import njit
from numba import jit
@jit
def sieve12(upToNumm: int=100000000, pnprimorial:int=3) -> list[int]:
# early in the function similar to:
C0:int=0
modprimorialdo:list[int]=[1,5]
lmodprimorialdo:int=len(modprimorialdo)
ddocol:int=C0
fdorowcol:list[int]=[C0,C0]
artp:int=C0
# and later:
# there are assignment statements for useful values for these variables, and then:
artp:int=int(ddocol+fdorowcol[C0]*lmodprimorialdo))
```
The full error report is :
```
Traceback (most recent call last):
File "/home/dakra/./primes2025a.py", line 810, in <module>
print("sieve12",n,pr,len(primesl:=sieve12(n,pr)), primesl[:10], primesl[-10:])
^^^^^^^^^^^^^
File "/home/dakra/.local/lib/python3.12/site-packages/numba/core/dispatcher.py", line 423, in _compile_for_args
error_rewrite(e, 'typing')
File "/home/dakra/.local/lib/python3.12/site-packages/numba/core/dispatcher.py", line 364, in error_rewrite
raise e.with_traceback(None)
numba.core.errors.TypingError: Failed in nopython mode pipeline (step: nopython frontend)
Type of variable 'artp.2' cannot be determined, operation: call $1078load_global.2($binop_add1104.10, func=$1078load_global.2, args=[Var($binop_add1104.10, primes2025a.py:240)], kws=(), vararg=None, varkwarg=None, target=None), location: /home/dakra/./primes2025a.py (240)
File "primes2025a.py", line 240:
def sieve12(upToNumm: int=100000000, pnprimorial:int=3)->list[int]:
<source elided>
artp:int=int(ddocol+fdorowcol[C0]*lmodprimorialdo)
^
```
In another case, the jit processing complained about one of the parameters.
What does it take to get the jit to recognize the explicit type specifications?
### CPython versions tested on:
3.12
### Operating systems tested on:
Linux
|
open
|
2025-01-06T15:58:31Z
|
2025-02-14T01:57:27Z
|
https://github.com/numba/numba/issues/9886
|
[
"more info needed",
"stale"
] |
dakra137
| 3
|
pydantic/pydantic
|
pydantic
| 10,443
|
Schema generation error when serialization schema holds a reference
|
### Initial Checks
- [X] I confirm that I'm using Pydantic V2
### Description
The following code currently raises:
```python
from typing import Annotated
from pydantic import BaseModel, PlainSerializer
class Sub(BaseModel):
pass
class Model(BaseModel):
sub: Annotated[
Sub,
PlainSerializer(lambda v: v, return_type=Sub),
]
# pydantic_core._pydantic_core.SchemaError: Definitions error: definition `__main__.Sub:97954749220704` was never filled
```
The reason is that, before schema cleaning, the core schema looks like:
```python
{
│ 'type': 'definitions',
│ 'schema': {'type': 'definition-ref', 'schema_ref': '__main__.Model:107773130038816'},
│ 'definitions': [
│ │ {
│ │ │ 'type': 'model',
│ │ │ 'cls': <class '__main__.Sub'>,
│ │ │ 'schema': {'type': 'model-fields', 'fields': {}, 'model_name': 'Sub', 'computed_fields': []},
│ │ │ 'config': {'title': 'Sub'},
│ │ │ 'ref': '__main__.Sub:107773128271744',
│ │ │ 'metadata': {'<stripped>'}
│ │ },
│ │ {
│ │ │ 'type': 'model',
│ │ │ 'cls': <class '__main__.Model'>,
│ │ │ 'schema': {
│ │ │ │ 'type': 'model-fields',
│ │ │ │ 'fields': {
│ │ │ │ │ 'sub': {
│ │ │ │ │ │ 'type': 'model-field',
│ │ │ │ │ │ 'schema': {
│ │ │ │ │ │ │ 'type': 'definition-ref',
│ │ │ │ │ │ │ 'schema_ref': '__main__.Sub:107773128271744',
│ │ │ │ │ │ │ 'serialization': {
│ │ │ │ │ │ │ │ 'type': 'function-plain',
│ │ │ │ │ │ │ │ 'function': <function Model.<lambda> at 0x7ae519b4f060>,
│ │ │ │ │ │ │ │ 'info_arg': False,
│ │ │ │ │ │ │ │ 'return_schema': {'type': 'definition-ref', 'schema_ref': '__main__.Sub:107773128271744'}
│ │ │ │ │ │ │ }
│ │ │ │ │ │ },
│ │ │ │ │ │ 'metadata': {'<stripped>'}
│ │ │ │ │ }
│ │ │ │ },
│ │ │ │ 'model_name': 'Model',
│ │ │ │ 'computed_fields': []
│ │ │ },
│ │ │ 'config': {'title': 'Model'},
│ │ │ 'metadata': {'<stripped>'},
│ │ │ 'ref': '__main__.Model:107773130038816'
│ │ }
│ ]
}
```
Which is quite unusual: the `'definition-ref'` schema to `Sub` has a `serialization` key. Pydantic does not expect this to happen, especially during schema simplification (when counting refs):
https://github.com/pydantic/pydantic/blob/01daafaab0aae71d2fb57c42461b3d021a3c56d4/pydantic/_internal/_core_utils.py#L447-L463
At some point, `count_refs` is called with the following schema:
```python
{
│ 'type': 'definition-ref',
│ 'schema_ref': '__main__.Sub:106278832701040',
│ 'serialization': {
│ │ 'type': 'function-plain',
│ │ 'function': <function Model.<lambda> at 0x7aed4dc920c0>,
│ │ 'info_arg': False,
│ │ 'return_schema': {'type': 'definition-ref', 'schema_ref': '__main__.Sub:106278832701040'}
│ }
}
```
the "top level" `__main__.Sub` ref is properly counted, but we skip recursion of the `serialization` schema. In the end, the ref counts for `__main__.Sub` is incorrect, and ends up incorrectly being inlined, which results in the schema validation error.
### Example Code
_No response_
### Python, Pydantic & OS Version
```Text
2.9
```
|
closed
|
2024-09-19T07:32:28Z
|
2024-09-19T13:42:42Z
|
https://github.com/pydantic/pydantic/issues/10443
|
[
"bug V2"
] |
Viicos
| 0
|
microsoft/nni
|
deep-learning
| 5,224
|
AttributeError: 'torch._C.Node' object has no attribute 'schema'
|
I use the tool to try to prue my model such as (https://github.com/microsoft/nni/blob/dab51f799f77aa72c18774faffaedf8d0ee2c977/examples/model_compress/pruning/admm_pruning_torch.py)
I only change the model(with restnet) and the dataloader:
But now there is some problem when I use ModelSpeedup
File "<ipython-input-7-25297990bbbb>", line 1, in <module>
ModelSpeedup(model, torch.randn([1, 2, 224, 224]).to(device), masks).speedup_model()
File "D:\anaconda3\lib\site-packages\nni\compression\pytorch\speedup\compressor.py", line 543, in speedup_model
self.infer_modules_masks()
File "D:\anaconda3\lib\site-packages\nni\compression\pytorch\speedup\compressor.py", line 380, in infer_modules_masks
self.update_direct_sparsity(curnode)
File "D:\anaconda3\lib\site-packages\nni\compression\pytorch\speedup\compressor.py", line 228, in update_direct_sparsity
func = jit_to_python_function(node, self)
File "D:\anaconda3\lib\site-packages\nni\compression\pytorch\speedup\jit_translate.py", line 554, in jit_to_python_function
return trans_func_dict[node.op_type](node, speedup)
File "D:\anaconda3\lib\site-packages\nni\compression\pytorch\speedup\jit_translate.py", line 488, in generate_aten_to_python
schema = c_node.schema()
AttributeError: 'torch._C.Node' object has no attribute 'schema'
|
closed
|
2022-11-11T14:10:09Z
|
2022-12-05T02:38:01Z
|
https://github.com/microsoft/nni/issues/5224
|
[] |
sunpeil
| 3
|
huggingface/transformers
|
pytorch
| 36,025
|
HIGGS Quantization not working properly
|
### System Info
**Environment**
```
- `transformers` version: 4.48.2
- Platform: Linux-5.4.210-39.1.pagevecsize-x86_64-with-glibc2.27
- Python version: 3.11.10
- Huggingface_hub version: 0.26.2
- Safetensors version: 0.4.5
- Accelerate version: 1.1.1
- Accelerate config: not found
- PyTorch version (GPU?): 2.4.0+cu121 (True)
- Tensorflow version (GPU?): not installed (NA)
- Flax version (CPU?/GPU?/TPU?): not installed (NA)
- Jax version: not installed
- JaxLib version: not installed
- Using distributed or parallel set-up in script?: <fill in>
- Using GPU in script?: <fill in>
- GPU type: NVIDIA A100-SXM4-80GB
- fast_hadamard_transform 1.0.4.post1
```
### Who can help?
@BlackSamorez
@SunMarc
@ArthurZucker
### Information
- [ ] The official example scripts
- [x] My own modified scripts
### Tasks
- [ ] An officially supported task in the `examples` folder (such as GLUE/SQuAD, ...)
- [ ] My own task or dataset (give details below)
### Reproduction
Recently, in the [PR](https://github.com/huggingface/transformers/pull/34997) HIGGS quantization from the paper [Pushing the Limits of Large Language Model Quantization via the Linearity Theorem](https://arxiv.org/abs/2411.17525) was introduced.
But when attempting to load the quantized `Llama-3.1-8B-Instruct `model in this format as follows:
```python
model_name = "meta-llama/Llama-3.1-8B-Instruct"
quantization_config = HiggsConfig(bits=4, p=2)
model = AutoModelForCausalLM.from_pretrained(
model_name,
device_map="auto",
torch_dtype=torch.bfloat16,
low_cpu_mem_usage=True,
quantization_config=quantization_config
)
model.config.use_cache = False
```
And doing forward pass with dummy inputs
```python
inputs = torch.randint(0, model.config.vocab_size, device="cuda", size=(8,))
with torch.no_grad():
outputs = model(inputs)
```
I get the following error in the RoPE:
```bash
File ~/miniconda3/envs/llm/lib/python3.11/site-packages/transformers/models/llama/modeling_llama.py:271, in LlamaAttention.forward(self, hidden_states, position_embeddings, attention_mask, past_key_value, cache_position, **kwargs)
[268](https://vscode-remote+ssh-002dremote-002bultramar.vscode-resource.vscode-cdn.net/home/dkuznedelev/FLUTE_Playground/~/miniconda3/envs/llm/lib/python3.11/site-packages/transformers/models/llama/modeling_llama.py:268) value_states = self.v_proj(hidden_states).view(hidden_shape).transpose(1, 2)
[270](https://vscode-remote+ssh-002dremote-002bultramar.vscode-resource.vscode-cdn.net/home/dkuznedelev/FLUTE_Playground/~/miniconda3/envs/llm/lib/python3.11/site-packages/transformers/models/llama/modeling_llama.py:270) cos, sin = position_embeddings
--> [271](https://vscode-remote+ssh-002dremote-002bultramar.vscode-resource.vscode-cdn.net/home/dkuznedelev/FLUTE_Playground/~/miniconda3/envs/llm/lib/python3.11/site-packages/transformers/models/llama/modeling_llama.py:271) query_states, key_states = apply_rotary_pos_emb(query_states, key_states, cos, sin)
[273](https://vscode-remote+ssh-002dremote-002bultramar.vscode-resource.vscode-cdn.net/home/dkuznedelev/FLUTE_Playground/~/miniconda3/envs/llm/lib/python3.11/site-packages/transformers/models/llama/modeling_llama.py:273) if past_key_value is not None:
[274](https://vscode-remote+ssh-002dremote-002bultramar.vscode-resource.vscode-cdn.net/home/dkuznedelev/FLUTE_Playground/~/miniconda3/envs/llm/lib/python3.11/site-packages/transformers/models/llama/modeling_llama.py:274) # sin and cos are specific to RoPE models; cache_position needed for the static cache
[275](https://vscode-remote+ssh-002dremote-002bultramar.vscode-resource.vscode-cdn.net/home/dkuznedelev/FLUTE_Playground/~/miniconda3/envs/llm/lib/python3.11/site-packages/transformers/models/llama/modeling_llama.py:275) cache_kwargs = {"sin": sin, "cos": cos, "cache_position": cache_position}
File ~/miniconda3/envs/llm/lib/python3.11/site-packages/transformers/models/llama/modeling_llama.py:169, in apply_rotary_pos_emb(q, k, cos, sin, position_ids, unsqueeze_dim)
[167](https://vscode-remote+ssh-002dremote-002bultramar.vscode-resource.vscode-cdn.net/home/dkuznedelev/FLUTE_Playground/~/miniconda3/envs/llm/lib/python3.11/site-packages/transformers/models/llama/modeling_llama.py:167) cos = cos.unsqueeze(unsqueeze_dim)
[168](https://vscode-remote+ssh-002dremote-002bultramar.vscode-resource.vscode-cdn.net/home/dkuznedelev/FLUTE_Playground/~/miniconda3/envs/llm/lib/python3.11/site-packages/transformers/models/llama/modeling_llama.py:168) sin = sin.unsqueeze(unsqueeze_dim)
--> [169](https://vscode-remote+ssh-002dremote-002bultramar.vscode-resource.vscode-cdn.net/home/dkuznedelev/FLUTE_Playground/~/miniconda3/envs/llm/lib/python3.11/site-packages/transformers/models/llama/modeling_llama.py:169) q_embed = (q * cos) + (rotate_half(q) * sin)
[170](https://vscode-remote+ssh-002dremote-002bultramar.vscode-resource.vscode-cdn.net/home/dkuznedelev/FLUTE_Playground/~/miniconda3/envs/llm/lib/python3.11/site-packages/transformers/models/llama/modeling_llama.py:170) k_embed = (k * cos) + (rotate_half(k) * sin)
[171](https://vscode-remote+ssh-002dremote-002bultramar.vscode-resource.vscode-cdn.net/home/dkuznedelev/FLUTE_Playground/~/miniconda3/envs/llm/lib/python3.11/site-packages/transformers/models/llama/modeling_llama.py:171) return q_embed, k_embed
RuntimeError: The size of tensor a (32) must match the size of tensor b (128) at non-singleton dimension 3
```
### Expected behavior
I would expect successful forward pass through the quantized model.
|
closed
|
2025-02-04T08:55:00Z
|
2025-02-19T05:35:52Z
|
https://github.com/huggingface/transformers/issues/36025
|
[
"bug"
] |
Godofnothing
| 3
|
ipython/ipython
|
jupyter
| 14,377
|
Not properly detecting IPython usage inside virtualenvs created via `uv venv`
|
I used the [`uv`](https://pypi.org/project/uv/) tool to create my venv and also to install ipython into it (via `uv pip install ipython`). When starting ipython I see the following warning:
```
C:\Users\jburnett1\Code\coordinates data analysis\.venv\Lib\site-packages\IPython\core\interactiveshell.py:937: UserWarning: Attempting to work in a virtualenv. If you encounter problems, please install IPython inside the virtualenv.
warn(
```
But if I examine the location of the IPython module it's definitely inside the currently activated venv:
```
import IPython
IPython
Out[4]: <module 'IPython' from 'C:\\Users\\jburnett1\\Code\\coordinates data analysis\\.venv\\Lib\\site-packages\\IPython\\__init__.py'>
import sys
sys.executable
Out[6]: 'C:\\Users\\jburnett1\\Code\\coordinates data analysis\\.venv\\Scripts\\python.exe'
```
I did find [this very old issue](https://github.com/ipython/ipython/issues/10955) which seemed to have a similar root cause, but the fix doesn't apparently work with the way that `uv` creates venvs & installs packages into them.
My version info:
```
sys.version
Out[7]: '3.12.1 (tags/v3.12.1:2305ca5, Dec 7 2023, 22:03:25) [MSC v.1937 64 bit (AMD64)]'
IPython.__version__
Out[8]: '8.22.2'
```
This is w/ uv version 0.1.6.
|
open
|
2024-03-28T14:18:37Z
|
2024-07-26T07:26:44Z
|
https://github.com/ipython/ipython/issues/14377
|
[] |
joshburnett
| 2
|
coqui-ai/TTS
|
deep-learning
| 2,649
|
[Bug]
|
### Describe the bug
Cannot find speaker file when using local nl models
### To Reproduce
1. download the nl model files
2. Run the code:
```
tts = TTS(model_path = <"model_path">,
config_path =<"config_path">,
vocoder_path = None,
vocoder_config_path = None,
progress_bar = False,
gpu = False)
```
### Expected behavior
The same normal behaviour as running `TTS('tts_models/nl/css10/vits', progress_bar=False, gpu=False)`
### Logs
```shell
FileNotFoundError: [Errno 2] No such file or directory: '/root/.cache/huggingface/hub/models--neongeckocom--tts-vits-css10-nl/snapshots/be7a7c7bee463588626b10777d7fc14ed8c07a3e/speaker_ids.json'
```
### Environment
```shell
TTS==0.13.3
```
### Additional context
working on a docker container
|
closed
|
2023-06-02T15:26:29Z
|
2023-06-05T08:00:42Z
|
https://github.com/coqui-ai/TTS/issues/2649
|
[
"bug"
] |
SophieDC98
| 1
|
pallets/flask
|
flask
| 5,004
|
Flask routes to return domain/sub-domains information
|
Currently when checking **flask routes** it provides all routes but **it is no way to see which routes are assigned to which subdomain**.
**Default server name:**
SERVER_NAME: 'test.local'
**Domains (sub-domains):**
test.test.local
admin.test.local
test.local
**Adding blueprints:**
app.register_blueprint(admin_blueprint,url_prefix='',subdomain='admin')
app.register_blueprint(test_subdomain_blueprint,url_prefix='',subdomain='test')
```
$ flask routes
* Tip: There are .env or .flaskenv files present. Do "pip install python-dotenv" to use them.
Endpoint Methods Rule
------------------------------------------------------- --------- ------------------------------------------------
admin_blueprint.home GET /home
test_subdomain_blueprint.home GET /home
static GET /static/<path:filename>
...
```
**Feature request**
It will be good to see something like below (that will make more clear which route for which subdomain, because now need to go and check configuration).
**If it is not possible to fix routes**, can you add or tell which method(s) should be used to get below information from flask?
```
$ flask routes
* Tip: There are .env or .flaskenv files present. Do "pip install python-dotenv" to use them.
Domain Endpoint Methods Rule
----------------- ---------------------------------------------------- ---------- ------------------------------------------------
admin.test.local admin_blueprint.home GET /home
test.test.local test_subdomain_blueprint.home GET /home
test.local static GET /static/<path:filename>
...
```
|
closed
|
2023-02-26T17:25:08Z
|
2023-04-29T00:05:02Z
|
https://github.com/pallets/flask/issues/5004
|
[
"cli"
] |
rimvislt
| 0
|
gradio-app/gradio
|
python
| 9,959
|
gr.File should add .html extension to URLs if possible
|
### Describe the bug
When using gr.File, the file uploaded keeps its extension. However, when using a URL like `https://liquipedia.net/starcraft2/Adept`, the file saved in the cache has no extension. This is technically correct. However, since it's HTML being downloaded, it should really save with the `.html` extension (equivalent to right-click and Save As in the browser). This makes it easier for a user to identify where it came from and also makes it easier for file type detection (avoiding the need for things like libmagic). Maybe it could just be a "default_web_extension" option that the user sets in the control.
The data source is lost when the upload happens (probably for security reasons?), so once a file has been downloaded to cache, you can't tell what it was originally or where it came from. The workaround would be to assume no extension means HTML, but that's a little error prone too.
### Have you searched existing issues? 🔎
- [X] I have searched and found no existing issues
### Reproduction
Just use gr.File to upload a URL by pasting a URL into the File Explorer pop-up that appears when clicking Upload.
### Screenshot
_No response_
### Logs
_No response_
### System Info
```shell
gradio==5.4.0
```
### Severity
I can work around it
|
closed
|
2024-11-14T14:10:07Z
|
2024-12-25T20:46:28Z
|
https://github.com/gradio-app/gradio/issues/9959
|
[
"enhancement",
"needs designing"
] |
JohnDuncanScott
| 2
|
CorentinJ/Real-Time-Voice-Cloning
|
deep-learning
| 747
|
PATH ISSUE module note recognized after installation
|
Hello I am so far doing good with the installation but finally when ready to start it, I receive the error
D:\Real Time Voice Cloning - Copy\Real-Time-Voice-Cloning-master>python demo_toolbox.py
Traceback (most recent call last):
File "demo_toolbox.py", line 2, in <module>
from toolbox import Toolbox
File "D:\Real Time Voice Cloning - Copy\Real-Time-Voice-Cloning-master\toolbox\__init__.py", line 1, in <module>
from toolbox.ui import UI
File "D:\Real Time Voice Cloning - Copy\Real-Time-Voice-Cloning-master\toolbox\ui.py", line 6, in <module>
from encoder.inference import plot_embedding_as_heatmap
File "D:\Real Time Voice Cloning - Copy\Real-Time-Voice-Cloning-master\encoder\inference.py", line 3, in <module>
from encoder.audio import preprocess_wav # We want to expose this function from here
File "D:\Real Time Voice Cloning - Copy\Real-Time-Voice-Cloning-master\encoder\audio.py", line 7, in <module>
import librosa
ModuleNotFoundError: No module named 'librosa'
The strange thing is that when prompted for librosa or to install librosa (specifically I perform a pip install) librosa is already installed. I believe this is simply a path issue because I have a dual hard drive setup. How exactly do I go about pathing the file to recognize librosa since the demo toolbox simply can't find the librosa. If possible can you maybe use imagery since I don't want to demand too much of your time, especially since you developed this amazing piece of software. Thank you for your diligence as well as pro level programming.
|
closed
|
2021-04-28T21:53:03Z
|
2021-05-04T17:20:36Z
|
https://github.com/CorentinJ/Real-Time-Voice-Cloning/issues/747
|
[] |
PsychoJack88
| 1
|
miguelgrinberg/python-socketio
|
asyncio
| 558
|
unable to stream saved videos from client to server
|
I can send my live video feed from client to server but when I am streaming the saved video stream I am unable to send it. It shows a segmentation fault.
Given below is my code
```python
from flask import Flask, render_template, request
import socketio
from time import sleep
import cv2
import json
import base64
cap=cv2.VideoCapture('768x576.avi')
sio = socketio.Client(engineio_logger=True)
@sio.event
def connect():
print("CONNECTED")
@sio.event
def send_data():
i=0
while(1):
ret,img=cap.read()
if ret:
img = cv2.resize(img, (0,0), fx=0.3, fy=0.3)
frame = cv2.imencode('.jpg', img)[1].tobytes()
frame = base64.encodebytes(frame).decode("utf-8")
message(frame)
else:
break
sleep(0.1)
def message(json):
print("/////////////////////////////500")
sio.emit('send',json)
@sio.event
def disconnect():
print("DISCONNECTED")
if __name__ == '__main__':
sio.connect('http://localhost:5000')
sio.wait()
```
Thanks in advance
|
closed
|
2020-11-02T13:14:02Z
|
2020-11-03T05:19:26Z
|
https://github.com/miguelgrinberg/python-socketio/issues/558
|
[
"question"
] |
AhmedBhati
| 2
|
dmlc/gluon-nlp
|
numpy
| 1,094
|
Add length normalized loss metrics in API
|
## Description
In typical machine translation tasks, training/validation metric loss is computed using some loss function normalized by the length of target sequence. For example, in `train_gnmt.py`, the metric is computed with the following code:
```python
loss = loss_function(out, tgt_seq[:, 1:], tgt_valid_length - 1).mean()
loss = loss * (tgt_seq.shape[1] - 1) / (tgt_valid_length - 1).mean()
```
Current Mxnet `metric.loss` does not support length normalization. It will be great to add length normalized metric in the api.
## References
- https://github.com/apache/incubator-mxnet/blob/master/python/mxnet/metric.py#L1661
- https://github.com/dmlc/gluon-nlp/blob/master/scripts/machine_translation/train_gnmt.py
|
closed
|
2020-01-06T07:18:17Z
|
2020-01-22T06:55:49Z
|
https://github.com/dmlc/gluon-nlp/issues/1094
|
[
"enhancement"
] |
liuzh47
| 0
|
napari/napari
|
numpy
| 7,329
|
napari freeze - QXcbIntegration: Cannot create platform OpenGL context, neither GLX nor EGL are enabled
|
### 🐛 Bug Report
when starting napari I get a frozen napari window displaying the terminal and the following error :
```
(napari) biop@c79840740ddb:~$ napari
WARNING: QStandardPaths: XDG_RUNTIME_DIR not set, defaulting to '/tmp/runtime-biop'
08:49:29 : WARNING : MainThread : QStandardPaths: XDG_RUNTIME_DIR not set, defaulting to '/tmp/runtime-biop'
WARNING: QXcbIntegration: Cannot create platform OpenGL context, neither GLX nor EGL are enabled
08:49:29 : WARNING : MainThread : QXcbIntegration: Cannot create platform OpenGL context, neither GLX nor EGL are enabled
WARNING: QXcbIntegration: Cannot create platform offscreen surface, neither GLX nor EGL are enabled
08:49:29 : WARNING : MainThread : QXcbIntegration: Cannot create platform offscreen surface, neither GLX nor EGL are enabled
WARNING: QXcbIntegration: Cannot create platform OpenGL context, neither GLX nor EGL are enabled
08:49:32 : WARNING : MainThread : QXcbIntegration: Cannot create platform OpenGL context, neither GLX nor EGL are enabled
WARNING: QXcbIntegration: Cannot create platform OpenGL context, neither GLX nor EGL are enabled
08:49:32 : WARNING : MainThread : QXcbIntegration: Cannot create platform OpenGL context, neither GLX nor EGL are enabled
WARNING: composeAndFlush: QOpenGLContext creation failed
```
### 💡 Steps to Reproduce
1. build a docker image with the docker file :
```dockerfile
ARG ALIAS=biop/
ARG BASE_IMAGE=0.1.0
FROM ${ALIAS}biop-vnc-base:${BASE_IMAGE}
USER root
# create napari env
RUN conda create --name napari napari=0.5.4 pyqt -c conda-forge -y
RUN chown -R biop:biop /home/biop/ \
&& chmod -R a+rwx /home/biop/
#################################################################
# Container start
USER biop
WORKDIR /home/biop
ENTRYPOINT ["/usr/local/bin/jupyter"]
CMD ["lab", "--allow-root", "--ip=*", "--port=8888", "--no-browser", "--NotebookApp.token=''", "--NotebookApp.allow_origin='*'", "--notebook-dir=/home/biop"]
```
2. start the image, follow the link to http://localhost:8888/lab
3. click on VNC icon , to start the desktop
4. start a terminal
5. type `source activate napari`
6. type napari
7. get the error
### 💡 Expected Behavior
napari window should pop-up normally
### 🌎 Environment
not working env : [napari_notworking.txt](https://github.com/user-attachments/files/17343548/napari_notworking.txt)
working env old version (0.4.18) of napari with devbio-napari [devbio_working.txt](https://github.com/user-attachments/files/17343551/devbio_working.txt)
### 💡 Additional Context
Thank you for any suggestion you might have
Cheers,
Romain
|
closed
|
2024-10-11T13:55:28Z
|
2025-03-10T09:54:12Z
|
https://github.com/napari/napari/issues/7329
|
[
"bug"
] |
romainGuiet
| 10
|
sunscrapers/djoser
|
rest-api
| 243
|
Changing Email Templates and Functionality
|
I was unable to change the email templates for Activation, Confirmation, or PasswordReset email. I wanted to add more to the context like a url with a url scheme for an app (not just http:// or https://).
I created a pull request #242 but if there is another way to do this currently, it would be much appreciated. I prefer to stay on the release branch because #242 maybe rejected or take a while to be made into a release.
|
closed
|
2017-11-01T13:13:22Z
|
2017-11-05T01:41:30Z
|
https://github.com/sunscrapers/djoser/issues/243
|
[] |
hammadzz
| 4
|
globaleaks/globaleaks-whistleblowing-software
|
sqlalchemy
| 3,185
|
Add support for Ubuntu Jammy Jellyfish (22.04)
|
This ticket is to extend support to [Ubuntu Jammy Jellyfish (22.04)](https://ubuntu.com/about/release-cycle) now in beta stage and for which release date is due to April 21, 2022.
|
closed
|
2022-02-28T14:43:44Z
|
2022-08-24T07:27:05Z
|
https://github.com/globaleaks/globaleaks-whistleblowing-software/issues/3185
|
[
"C: Packaging"
] |
evilaliv3
| 0
|
encode/uvicorn
|
asyncio
| 1,851
|
Route identification problem with pyinstaller
|
fastapi 0.89.1
uvicorn[standard] 0.20.0 (it did not happed on 0.17.6)
python 3.11.1
pyinstaller 5.7.0
This promble only happens when i use pyinstaller package the program with version 0.20.0,and when i reduce program version to 0.17.6,it fix.

When i use thwo routers like this,the router [name]with[s] will not get the state
router config/{name} can get state,but /configs cannot get the state,when i change /configs to /configlist , it works.
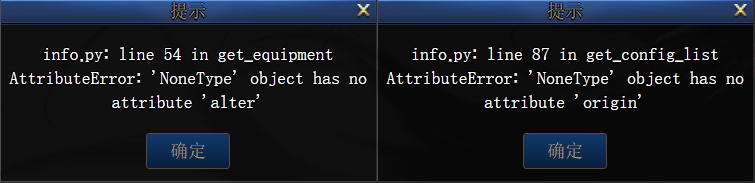
|
closed
|
2023-01-28T14:40:42Z
|
2023-01-28T21:05:42Z
|
https://github.com/encode/uvicorn/issues/1851
|
[] |
wxh0402
| 0
|
akfamily/akshare
|
data-science
| 5,620
|
AKShare 接口问题报告 | AKShare Interface Issue Report
|
> 欢迎加入《数据科学实战》知识星球,交流财经数据与量化投资相关内容 |
> Welcome to join "Data Science in Practice" Knowledge
> Community for discussions on financial data and quantitative investment.
>
> 详细信息参考 | For detailed information, please visit::https://akshare.akfamily.xyz/learn.html
## 前提 | Prerequisites
遇到任何问题,请先将您的 AKShare 版本升级到**最新版**,可以通过如下命令升级 | Before reporting any issues, please upgrade
your AKShare to the **latest version** using the following command::
```
pip install akshare --upgrade # Python 版本需要大于等于 3.8 | Python version requirement ≥ 3.8
```
## 如何提交问题 | How to Submit an Issue
提交问题的同时,请提交以下相关信息,以更精准的解决问题。| Please provide the following information when
submitting an issue for more accurate problem resolution.
**不符合提交规范的 issues 会被关闭!** | **Issues that don't follow these guidelines will be closed!**
**详细问题描述** | Detailed Problem Description
1. 请先详细阅读文档对应接口的使用方式 | Please read the documentation thoroughly for the
relevant interface:https://akshare.akfamily.xyz
2. 操作系统版本,目前只支持 64 位操作系统 | Operating system version (64-bit only supported)
3. Python 版本,目前只支持 3.8 以上的版本 | Python version (must be 3.8 or above)
4. AKShare 版本,请升级到最新版 | AKShare version (please upgrade to latest)
5. 接口的名称和相应的调用代码 | Interface name and corresponding code
6. 接口报错的截图或描述 | Screenshot or description of the error
7. 期望获得的正确结果 | Expected correct results
|
closed
|
2025-02-16T05:43:44Z
|
2025-02-16T09:32:27Z
|
https://github.com/akfamily/akshare/issues/5620
|
[
"bug"
] |
cnfaxian
| 1
|
vimalloc/flask-jwt-extended
|
flask
| 280
|
flask_restplus and setting cookies
|
I'm using `Flask_RESTPlus` and I'm trying to set JWT cookies but looks like `flask_restplus` don't allow to return a `jsonify object` and I can't set the cookies in the response.
```
@api.route('/sign-in')
class SignInResource(Resource):
def post(self):
username_or_email = request.json.get('username_or_email', None)
password = request.json.get('password', None)
if username_or_email != "test" and password != test:
return {"error": "bad credentials"}
access_token = create_access_token(identity=user.id)
refresh_token = create_refresh_token(identity=user.id)
ret = {'sign_in': True}
resp = jsonify(ret)
set_access_cookies(resp, access_token)
set_refresh_cookies(resp, refresh_token)
return resp, 200 # DONT ALLOW return JSONIFY Object
```
Is there a workaround for this?
|
closed
|
2019-10-17T07:59:22Z
|
2019-10-17T14:52:17Z
|
https://github.com/vimalloc/flask-jwt-extended/issues/280
|
[] |
psdon
| 2
|
zappa/Zappa
|
django
| 928
|
[Migrated] Is it possible to not include the slug in the function name?
|
Originally from: https://github.com/Miserlou/Zappa/issues/2194 by [buckmaxwell](https://github.com/buckmaxwell)
<!--- Provide a general summary of the issue in the Title above -->
## Context
Is it possible to not include the slug in the function name? Ie, change default function name from `<project-name>-<stage-name>` to just `<project-name>`.
I'd like to be able to deploy / update an existing lambda by supplying an arn or something. My lambda was created by terraform.
|
closed
|
2021-02-20T13:24:39Z
|
2024-04-13T19:36:45Z
|
https://github.com/zappa/Zappa/issues/928
|
[
"no-activity",
"auto-closed"
] |
jneves
| 2
|
yihong0618/running_page
|
data-visualization
| 558
|
设置`IGNORE_BEFORE_SAVING=1`后`gen_svg.py`报错
|
看了下代码,应该是 `run_page/gpxtrackposter/track.py`中`load_from_db`函数的问题。blame下,是在这个[commit](https://github.com/yihong0618/running_page/commit/f17a3e694a2ba102a218efc16e21a8cf83f0053b#diff-ea1ab7ec71fbeb34ad5d08163ac064c11b28e4ccbe97f75b1a876015aac51a8e)引入的。
未设置该环境变量时,不会触发bug。设置该变量,则导致`summary_polyline`发生未定义先引用错误。
按照我的理解,代码应该改成这个样子:
```python
if IGNORE_BEFORE_SAVING:
summary_polyline = filter_out(activity.summary_polyline)
else:
summary_polyline = activity.summary_polyline
```
|
closed
|
2023-12-02T13:31:06Z
|
2023-12-02T14:18:51Z
|
https://github.com/yihong0618/running_page/issues/558
|
[] |
conanyangqun
| 4
|
ivy-llc/ivy
|
pytorch
| 28,627
|
Fix Frontend Failing Test: tensorflow - activations.tensorflow.keras.activations.relu
|
To-do List: https://github.com/unifyai/ivy/issues/27499
|
closed
|
2024-03-17T23:57:00Z
|
2024-03-25T12:47:11Z
|
https://github.com/ivy-llc/ivy/issues/28627
|
[
"Sub Task"
] |
ZJay07
| 0
|
kizniche/Mycodo
|
automation
| 950
|
Python 3 code input not storing measurements after upgrading to 8.9.1
|
### Issue Summary
A Python 3 code input that was successfully running in Mycodo 8.8.8 without issues is now not storing measurements to the database. Mycodo is logging many of these errors:
`ERROR - mycodo.controllers.controller_input_957ddbe6 - StopIteration raised 3 times. Possibly could not read input. Ensure it's connected properly and detected.`
The code DOES successfully return the expected value, it just doesn't get stored. I can see the returned value in the Data -> Live view, and also on any of the dashboard widgets that use the live value. I can NOT view historical data (e.g. on either of the graph types) as I could on 8.8.8.
This is on a fresh install of Mycodo 8.9.1 with a single Python 3 input configured in the same manner (with the same code) that it was running successfully on 8.8.8.
### Versions:
- Mycodo Version: 8.9.1
- Raspberry Pi Version: 4
- Raspbian OS Version: Buster Lite 2020-02-13
### Reproducibility
The Python 3 code manipulates a JSN-SR04T sensor, so you'd need one to test. On a side note, I see that the latest version of Mycodo supports this sensor natively, but my environment has enough background noise the the sensor results are far too inconsistent to use without some manipulation, which I accomplish via code.
1. Go to 'Setup' -> 'Input'
2. Click on 'Linux: Python 3 Code: Store Value(s) [Mycodo]'
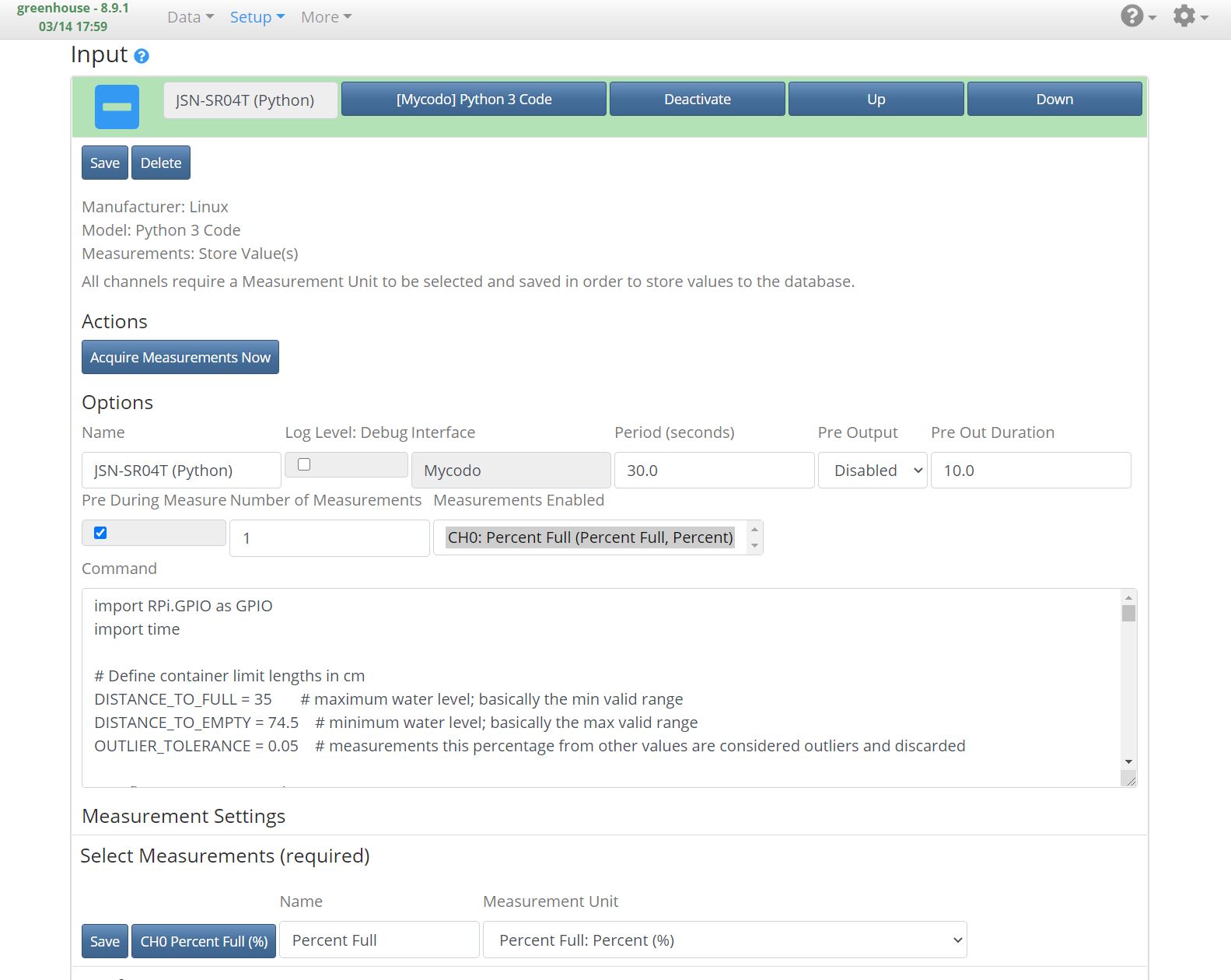
3. Configure as shown in this screenshot:
The full Python 3 code:
```
import RPi.GPIO as GPIO
import time
# Define container limit lengths in cm
DISTANCE_TO_FULL = 35 # maximum water level; basically the min valid range
DISTANCE_TO_EMPTY = 74.5 # minimum water level; basically the max valid range
OUTLIER_TOLERANCE = 0.05 # measurements this percentage from other values are considered outliers and discarded
# Define GPIO to use on Pi
GPIO_TRIGGER = 23
GPIO_ECHO = 18
TRIGGER_TIME = 0.00001
MAX_TIME = 0.1 # max time waiting for response in case something is missed
TIME_BETWEEN_MEASUREMENTS = 0.5 # time to sleep between each measurement
GPIO.setmode(GPIO.BCM)
GPIO.setwarnings(False)
GPIO.setup(GPIO_TRIGGER, GPIO.OUT) # Trigger
GPIO.setup(GPIO_ECHO, GPIO.IN, pull_up_down=GPIO.PUD_UP) # Echo
GPIO.output(GPIO_TRIGGER, False)
# This function measures a distance
def measure():
# Pulse the trigger/echo line to initiate a measurement
GPIO.output(GPIO_TRIGGER, True)
time.sleep(TRIGGER_TIME)
GPIO.output(GPIO_TRIGGER, False)
# ensure start time is set in case of very quick return
start = time.time()
timeout = start + MAX_TIME
# set line to input to check for start of echo response
while GPIO.input(GPIO_ECHO) == 0 and start <= timeout:
start = time.time()
if(start > timeout):
return -1
stop = time.time()
timeout = stop + MAX_TIME
# Wait for end of echo response
while GPIO.input(GPIO_ECHO) == 1 and stop <= timeout:
stop = time.time()
if(stop <= timeout):
elapsed = stop-start
distance = float(elapsed * 34300)/2.0
else:
return -1
return distance
# take 3 measurements, discard outliers, return averge result
def sample():
measurements = []
attempts = 0
while len(measurements) < 3 and attempts < 15:
if attempts > 0:
time.sleep(TIME_BETWEEN_MEASUREMENTS)
distance = measure()
attempts += 1
if(distance >= DISTANCE_TO_FULL and distance <= DISTANCE_TO_EMPTY):
measurements.append(distance)
if len(measurements) == 3:
# check for and remove outlier measurements
for i in range(len(measurements)):
if is_outlier(measurements, i):
measurements.pop(i)
# if there are at least 2 good data points remaining, take the average and return it
if len(measurements) >= 2:
measure_sum = 0
for i in range(len(measurements)):
measure_sum += measurements[i]
measure_avg = measure_sum/len(measurements)
return (measure_avg)
else:
# we didn't have at least 2 good data points
return -2
else:
return -1
# run the sample function twice, discard result if both values not within OUTLIER_TOLERANCE of each other
def double_sample():
trials = [-1, -1]
# run the two trials
for i in range(len(trials)):
attempt = 0
while attempt < 6 and trials[i] < 0:
attempt += 1
trials[i] = sample()
# compare the two results
if is_outlier(trials, 0):
# the two values aren't consistent
return -1
else:
trial_avg = (trials[0] + trials[1])/2
return trial_avg
# compare the value of the list item at the given index to every other item in the list
# if the given item varies by more than OUTLIER_TOLERANCE compared to the other items, it's an outlier
def is_outlier(list, index):
if index > len(list)-1:
return False
else:
outlier_compares = 0
outlier_true = 0
value = list[index]
for i in range(len(list)):
if i != index:
outlier_compares += 1
diff = abs(list[i] - value)
diff_percent = diff/list[i]
if diff_percent > OUTLIER_TOLERANCE:
outlier_true += 1
else:
break
if outlier_compares == outlier_true:
# every other item has a value that is off by at least OUTLIER_TOLERANCE, this is an outlier
return True
else:
# this item is within OUTLIER_TOLERANCE of every other item
return False
# take the measurement
attempts = 0
percent_full = -1
while attempts < 5:
attempts += 1
result = double_sample()
if result >= 0:
percent_full = (1-(result/DISTANCE_TO_EMPTY)) * 100
break
GPIO.cleanup()
# Store measurements in database (must specify the channel and measurement)
self.store_measurement(channel=0, measurement=percent_full)
```
4. Save/activate the input
### Expected behavior
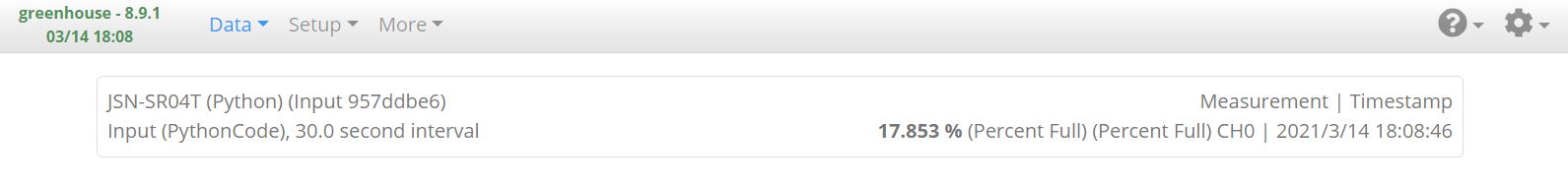
From here, I can navigate to Data -> Live and see the latest measurement as expected:
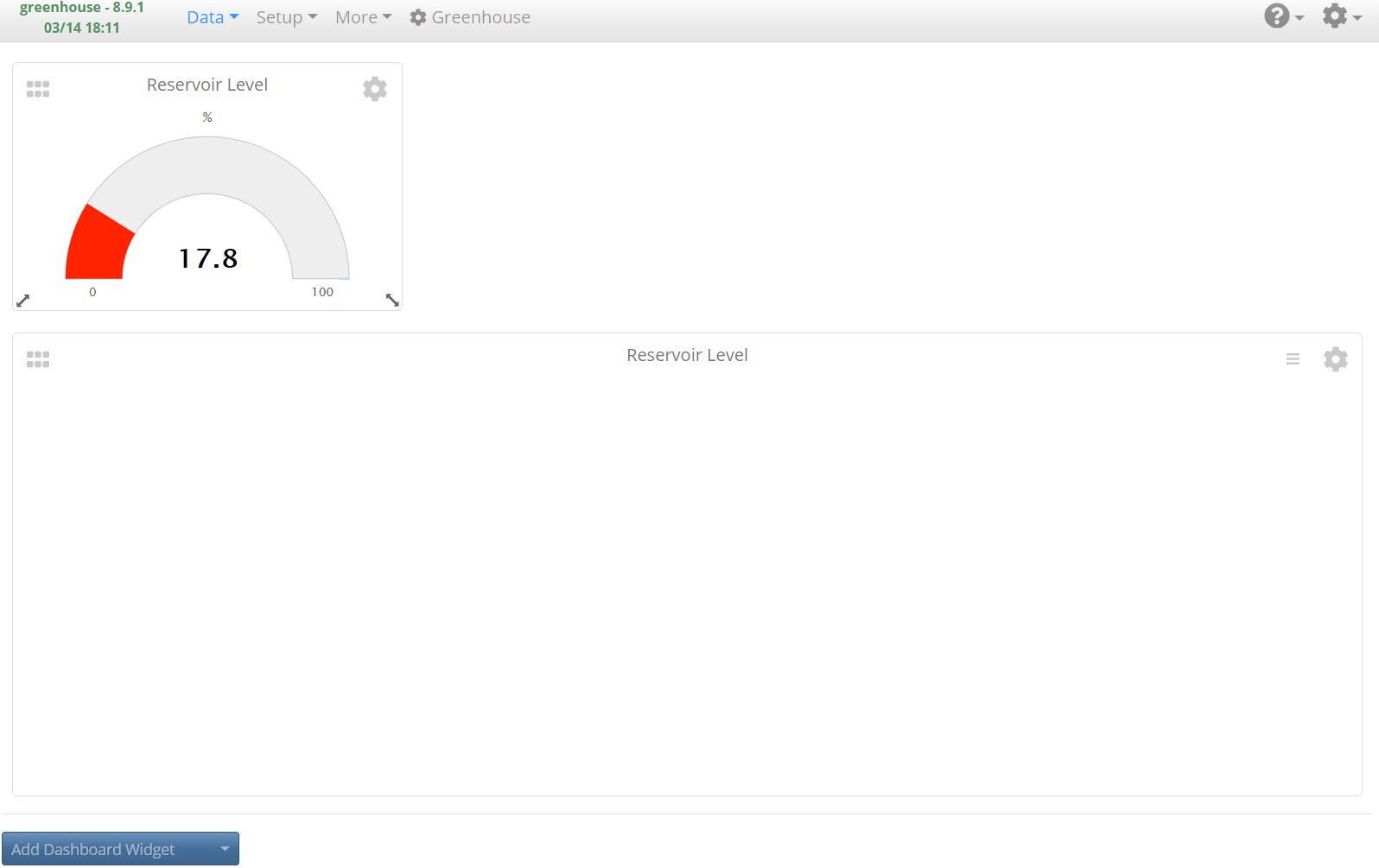
But if I try to create a graph widget, or view async graph data, they're just blank - presumably because historical data isn't being saved to the database properly:
Viewing the log, I see that a bunch of StopIteration errors are being thrown on many (but not all, judging by the timestamps) executions:
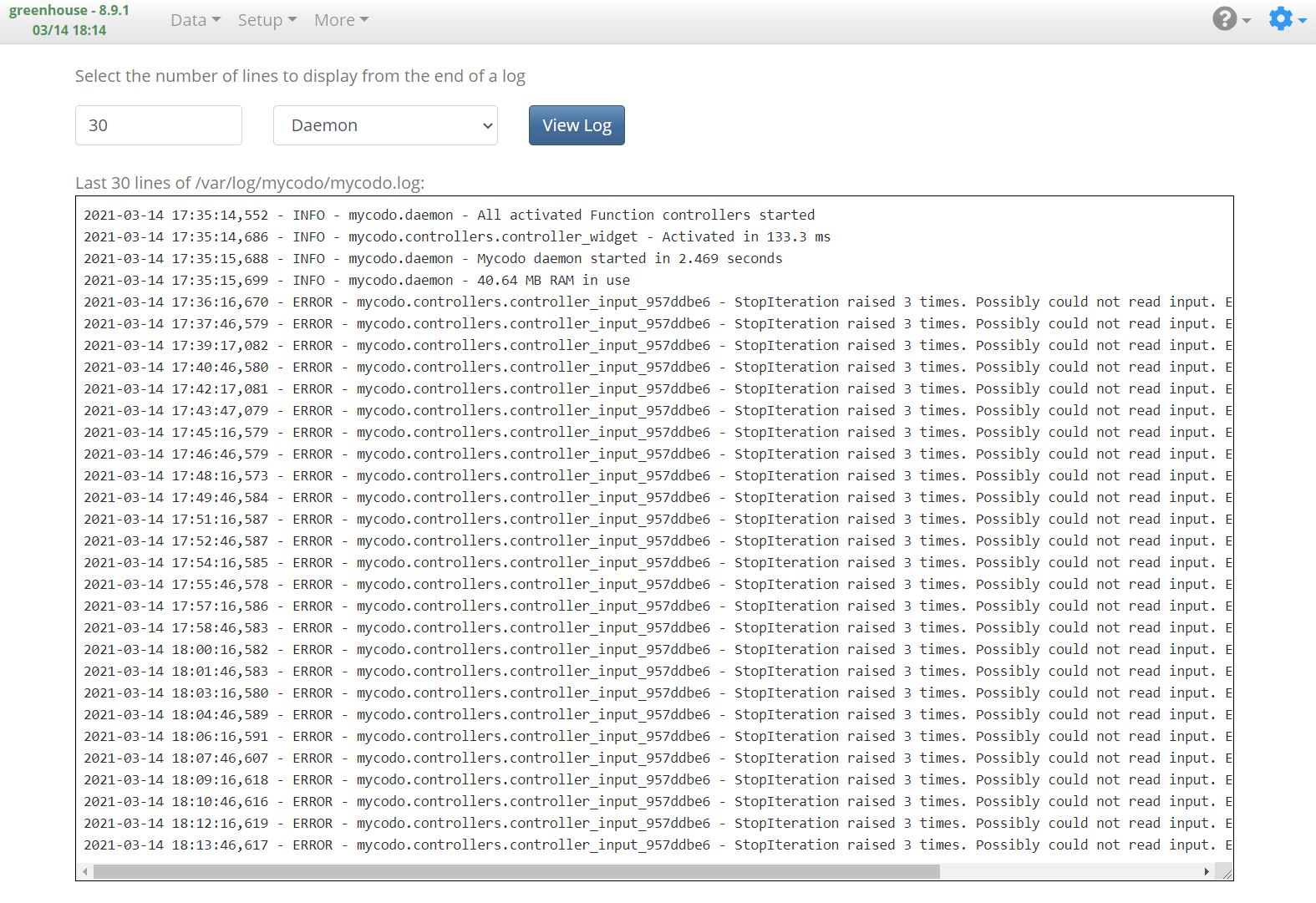
### Additional context
This code has been successfully running on a Mycodo 8.8.8 install for a couple weeks without issues. I attempted to use Mycodo's upgrade function to move to 8.9.1, but that broke my install (HTTP 500 errors on most pages, presumably from a dropped custom measurement I had created - I already created a [separate issue](https://github.com/kizniche/Mycodo/issues/949) for that). So I moved my Mycodo installation to a backup directory and installed 8.9.1 from scratch. Just mentioning in case that might play a part in the issue.
|
closed
|
2021-03-14T22:25:22Z
|
2021-03-18T17:01:36Z
|
https://github.com/kizniche/Mycodo/issues/950
|
[
"bug",
"Fixed and Committed"
] |
rbbrdckybk
| 6
|
jonaswinkler/paperless-ng
|
django
| 140
|
Show matching documents when clicking on document count
|
Hi :wave:,
I would like to make the following feature request proposal:
```bash
When I click on Tags in the Manage Menu
And I click on the document count of a specific tag
Then I see all associated documents in the document view using the tag as a filter
When I click on Correspondents in the Manage Menu
And I click on the document count of a specific correspondent
Then I see all associated documents in the document view using the correspondent as a filter
When I click on Document types in the Manage Menu
And I click on the document count of a specific document type
Then I see all associated documents in the document view using the document type as a filter
```
* Maybe this is obsolete with the new filter redesign, but I find it useful to reach the documents within these views
* I don't mind if the link is from document count, tag name, or else
|
closed
|
2020-12-15T16:30:54Z
|
2022-06-25T12:58:23Z
|
https://github.com/jonaswinkler/paperless-ng/issues/140
|
[
"feature request"
] |
Tooa
| 4
|
graphql-python/graphql-core
|
graphql
| 72
|
Exception instance returned from resolver is raised within executor instead of being passed down for further resolution.
|
Today when experimenting with 3rd party library for data validation (https://github.com/samuelcolvin/pydantic) I've noticed that when I take map of validation errors from it (which is dict of lists of `ValueError` and `TypeError` subclasses instances), I need to implement extra step to convert those errors to something else because query executor includes check for `isinstance(result, Exception)`. This check makes it raise returned exception instance, effectively short-circuiting further resolution:
https://github.com/graphql-python/graphql-core-next/blob/master/src/graphql/execution/execute.py#L731
The fix for issue was considerably simple to come up with: just write util that converts those errors to a dict as they are included in my result's `validation_errors` key, but I feel such boilerplate should be unnecessary:
```
try:
... run pydantic validation here
except (PydanticTypeError, PydanticValueError) as error:
return {"validation_errors": flatten_validation_error(error)}
```
Is this implementation a result of something in spec, or mechanism used to keep other feature's (eg. error propagation) code simple? I think we should consider supporting this use-case. Considerable number of libraries use exceptions for messaging (eg. Django with its `ValidationError` and bunch of `core.exceptions.*`)
|
closed
|
2019-12-14T22:35:16Z
|
2020-01-06T13:07:34Z
|
https://github.com/graphql-python/graphql-core/issues/72
|
[] |
rafalp
| 1
|
numpy/numpy
|
numpy
| 28,365
|
BUG: duplication in `ufunc.types`
|
### Describe the issue:
I'm working on type-test code generation, and I assumed that the `ufunc.types` entries would be unique. But as it turns out, that's not the case.
Specifically, these are all the duplicate `types` values in the public `numpy` namespace:
```
acos: e->e f->f d->d
acosh: e->e f->f d->d
asin: e->e f->f d->d
asinh: e->e f->f d->d
atan: e->e f->f d->d
atanh: e->e f->f d->d
cbrt: e->e f->f d->d
ceil: f->f d->d
cosh: e->e f->f d->d
exp: f->f d->d
exp2: e->e f->f d->d
expm1: e->e f->f d->d
floor: f->f d->d
log: f->f d->d
log10: e->e f->f d->d
log1p: e->e f->f d->d
log2: e->e f->f d->d
pow: ee->e ff->f dd->d
rint: f->f d->d
sinh: e->e f->f d->d
sqrt: f->f d->d
tan: e->e f->f d->d
tanh: e->e f->f d->d
trunc: f->f d->d
```
I'm guesing it's related to the e.g. `single` vs `float64` stuff.
In my use-case, it's not difficult to work around. But I thought maybe this could maybe cause some performance issues or something 🤷🏻.
### Reproduce the code example:
```python
import numpy as np
assert len(np.log.types) == len(set(np.log.types))
```
### Error message:
```shell
Traceback (most recent call last):
File "stop_eating_that_clay__son.py", line 3, in <module>
assert len(set(np.log.types)) == len(np.log.types)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
AssertionError
```
### Python and NumPy Versions:
```
2.2.3
3.13.2 (main, Feb 7 2025, 04:13:54) [GCC 11.4.0]
```
### Runtime Environment:
```
[{'numpy_version': '2.2.3',
'python': '3.13.2 (main, Feb 7 2025, 04:13:54) [GCC 11.4.0]',
'uname': uname_result(system='Linux', node='pop-os', release='6.9.3-76060903-generic', version='#202405300957~1738770968~22.04~d5f7c84 SMP PREEMPT_DYNAMIC Wed F', machine='x86_64')},
{'simd_extensions': {'baseline': ['SSE', 'SSE2', 'SSE3'],
'found': ['SSSE3',
'SSE41',
'POPCNT',
'SSE42',
'AVX',
'F16C',
'FMA3',
'AVX2'],
'not_found': ['AVX512F',
'AVX512CD',
'AVX512_KNL',
'AVX512_KNM',
'AVX512_SKX',
'AVX512_CLX',
'AVX512_CNL',
'AVX512_ICL']}},
{'architecture': 'Haswell',
'filepath': '/home/joren/.pyenv/versions/3.13.2/lib/python3.13/site-packages/numpy.libs/libscipy_openblas64_-6bb31eeb.so',
'internal_api': 'openblas',
'num_threads': 32,
'prefix': 'libscipy_openblas',
'threading_layer': 'pthreads',
'user_api': 'blas',
'version': '0.3.28'},
{'architecture': 'Haswell',
'filepath': '/home/joren/.pyenv/versions/3.13.2/lib/python3.13/site-packages/scipy.libs/libscipy_openblas-68440149.so',
'internal_api': 'openblas',
'num_threads': 32,
'prefix': 'libscipy_openblas',
'threading_layer': 'pthreads',
'user_api': 'blas',
'version': '0.3.28'}]
```
### Context for the issue:
_No response_
|
open
|
2025-02-20T18:39:58Z
|
2025-02-25T11:06:53Z
|
https://github.com/numpy/numpy/issues/28365
|
[
"00 - Bug"
] |
jorenham
| 1
|
MaartenGr/BERTopic
|
nlp
| 1,767
|
How to get more than 3 representative docs per topic via get_topic_info()?
|
Hey, thank you so much for making this library! Super awesome.
I've seen a bunch of issues here requesting this but haven't found a straightforward easy way to specify it via get_topic_info() as it contains a lot of the information I need. I wish there was a parameter in there like get_topic_info(number_of_representative_documents=3) that I could modify.
I'm not sure that `_extract_representative_docs` will work in my context as I'm using umap, hdbscan, and gpt for topic labels, no tfidf or anything, which seems to be a required parameter
|
open
|
2024-01-22T22:44:49Z
|
2024-03-20T09:09:16Z
|
https://github.com/MaartenGr/BERTopic/issues/1767
|
[] |
youssefabdelm
| 3
|
python-gitlab/python-gitlab
|
api
| 2,732
|
first run in jenkins always fails with GitlabHttpError: 403: insufficient_scope
|
## Description of the problem, including code/CLI snippet
we use jenkins for our builds and use semantic-versioning. usually, when someone pushed something into master/main, jenkins creates a new version, tags it to gitlab and builds the distribution. unfortunately, since a few weeks(not sure about that) we have a very wierd behaviour. the first run fails.
## Expected Behavior
a regular jenkins build with tags pushed to gitlab and no error
## Actual Behavior
we get an error GitlabHttpError: 403: insufficient_scope when jenkins runs the first time on that branch, when i then start it manually, it works. even more wierd: the tag is created!
```
(.pyenv-python) C:\Users\jenkinsSA\AppData\Local\Jenkins\.jenkins\workspace\usermgmtautomation_master>
git checkout master
Switched to branch 'master'
[Pipeline] bat
(.pyenv-python) C:\Users\jenkinsSA\AppData\Local\Jenkins\.jenkins\workspace\usermgmtautomation_master>
semantic-release version 1>version.txt
The next version is: 0.7.3! \U0001f680
No build command specified, skipping
[08:54:15] ERROR [semantic_release.cli.commands.version] version.py:553
ERROR version.version: 403:
insufficient_scope
+--- Traceback (most recent call last) ----+
| C:\Users\jenkinsSA\AppData\Local\Jenkins |
| \.jenkins\workspace\usermgmtautomation_m |
| aster\.pyenv-python\Lib\site-packages\gi |
| tlab\exceptions.py:336 in wrapped_f |
| |
| 333 @functools.wraps(f) |
| 334 def wrapped_f(*args: Any, |
| 335 try: |
| > 336 return f(*args, ** |
| 337 except GitlabHttpError |
| 338 raise error(e.erro |
| 339 |
| |
| C:\Users\jenkinsSA\AppData\Local\Jenkins |
| \.jenkins\workspace\usermgmtautomation_m |
| aster\.pyenv-python\Lib\site-packages\gi |
| tlab\mixins.py:300 in create |
| |
| 297 |
| 298 # Handle specific URL for |
| 299 path = kwargs.pop("path", |
| > 300 server_data = self.gitlab. |
| 301 if TYPE_CHECKING: |
| 302 assert not isinstance( |
| 303 assert self._obj_cls i |
| |
| C:\Users\jenkinsSA\AppData\Local\Jenkins |
| \.jenkins\workspace\usermgmtautomation_m |
| aster\.pyenv-python\Lib\site-packages\gi |
| tlab\client.py:1021 in http_post |
| |
| 1018 query_data = query_data o |
| 1019 post_data = post_data or |
| 1020 |
| > 1021 result = self.http_reques |
| 1022 "post", |
| 1023 path, |
| 1024 query_data=query_data |
| |
| C:\Users\jenkinsSA\AppData\Local\Jenkins |
| \.jenkins\workspace\usermgmtautomation_m |
| aster\.pyenv-python\Lib\site-packages\gi |
| tlab\client.py:794 in http_request |
| |
| 791 response_body |
| 792 ) |
| 793 |
| > 794 raise gitlab.exceptio |
| 795 response_code=res |
| 796 error_message=err |
| 797 response_body=res |
+------------------------------------------+
GitlabHttpError: 403: insufficient_scope
The above exception was the direct cause of
the following exception:
+--- Traceback (most recent call last) ----+
| C:\Users\jenkinsSA\AppData\Local\Jenkins |
| \.jenkins\workspace\usermgmtautomation_m |
| aster\.pyenv-python\Lib\site-packages\se |
| mantic_release\hvcs\gitlab.py:131 in |
| create_or_update_release |
| |
| 128 self, tag: str, release_no |
| 129 ) -> str: |
| 130 try: |
| > 131 return self.create_rel |
| 132 tag=tag, release_n |
| 133 ) |
| 134 except gitlab.GitlabCreate |
| |
| C:\Users\jenkinsSA\AppData\Local\Jenkins |
| \.jenkins\workspace\usermgmtautomation_m |
| aster\.pyenv-python\Lib\site-packages\se |
| mantic_release\helpers.py:52 in _wrapper |
| |
| 49 ) |
| 50 |
| 51 # Call function |
| > 52 result = func(*args, * |
| 53 |
| 54 # Log result |
| 55 logger.debug("%s -> %s |
| |
| C:\Users\jenkinsSA\AppData\Local\Jenkins |
| \.jenkins\workspace\usermgmtautomation_m |
| aster\.pyenv-python\Lib\site-packages\se |
| mantic_release\hvcs\gitlab.py:97 in |
| create_release |
| |
| 94 client.auth() |
| 95 log.info("Creating release |
| 96 # ref: https://docs.gitlab/ |
| > 97 client.projects.get(self.o |
| 98 { |
| 99 "name": "Release " |
| 100 "tag_name": tag, |
| |
| C:\Users\jenkinsSA\AppData\Local\Jenkins |
| \.jenkins\workspace\usermgmtautomation_m |
| aster\.pyenv-python\Lib\site-packages\gi |
| tlab\exceptions.py:338 in wrapped_f |
| |
| 335 try: |
| 336 return f(*args, ** |
| 337 except GitlabHttpError |
| > 338 raise error(e.erro |
| 339 |
| 340 return cast(__F, wrapped_f |
| 341 |
+------------------------------------------+
GitlabCreateError: 403: insufficient_scope
During handling of the above exception,
another exception occurred:
+--- Traceback (most recent call last) ----+
| C:\Users\jenkinsSA\AppData\Local\Jenkins |
| \.jenkins\workspace\usermgmtautomation_m |
| aster\.pyenv-python\Lib\site-packages\gi |
| tlab\exceptions.py:336 in wrapped_f |
| |
| 333 @functools.wraps(f) |
| 334 def wrapped_f(*args: Any, |
| 335 try: |
| > 336 return f(*args, ** |
| 337 except GitlabHttpError |
| 338 raise error(e.erro |
| 339 |
| |
| C:\Users\jenkinsSA\AppData\Local\Jenkins |
| \.jenkins\workspace\usermgmtautomation_m |
| aster\.pyenv-python\Lib\site-packages\gi |
| tlab\mixins.py:368 in update |
| |
| 365 ) |
| 366 |
| 367 http_method = self._get_up |
| > 368 result = http_method(path, |
| 369 if TYPE_CHECKING: |
| 370 assert not isinstance( |
| 371 return result |
| |
| C:\Users\jenkinsSA\AppData\Local\Jenkins |
| \.jenkins\workspace\usermgmtautomation_m |
| aster\.pyenv-python\Lib\site-packages\gi |
| tlab\client.py:1075 in http_put |
| |
| 1072 query_data = query_data o |
| 1073 post_data = post_data or |
| 1074 |
| > 1075 result = self.http_reques |
| 1076 "put", |
| 1077 path, |
| 1078 query_data=query_data |
| |
| C:\Users\jenkinsSA\AppData\Local\Jenkins |
| \.jenkins\workspace\usermgmtautomation_m |
| aster\.pyenv-python\Lib\site-packages\gi |
| tlab\client.py:794 in http_request |
| |
| 791 response_body |
| 792 ) |
| 793 |
| > 794 raise gitlab.exceptio |
| 795 response_code=res |
| 796 error_message=err |
| 797 response_body=res |
+------------------------------------------+
GitlabHttpError: 403: insufficient_scope
The above exception was the direct cause of
the following exception:
+--- Traceback (most recent call last) ----+
| C:\Users\jenkinsSA\AppData\Local\Jenkins |
| \.jenkins\workspace\usermgmtautomation_m |
| aster\.pyenv-python\Lib\site-packages\se |
| mantic_release\cli\commands\version.py:5 |
| 47 in version |
| |
| 544 noop_report(f"would ha |
| 545 else: |
| 546 try: |
| > 547 release_id = hvcs_ |
| 548 tag=new_versio |
| 549 release_notes= |
| 550 prerelease=new |
| |
| C:\Users\jenkinsSA\AppData\Local\Jenkins |
| \.jenkins\workspace\usermgmtautomation_m |
| aster\.pyenv-python\Lib\site-packages\se |
| mantic_release\helpers.py:52 in _wrapper |
| |
| 49 ) |
| 50 |
| 51 # Call function |
| > 52 result = func(*args, * |
| 53 |
| 54 # Log result |
| 55 logger.debug("%s -> %s |
| |
| C:\Users\jenkinsSA\AppData\Local\Jenkins |
| \.jenkins\workspace\usermgmtautomation_m |
| aster\.pyenv-python\Lib\site-packages\se |
| mantic_release\hvcs\gitlab.py:141 in |
| create_or_update_release |
| |
| 138 self.owner, |
| 139 self.repo_name, |
| 140 ) |
| > 141 return self.edit_relea |
| 142 |
| 143 def compare_url(self, from_rev |
| 144 return |
| f"[https://{self.hvcs_domain}/{self](https://{self.hvcs_domain}/%7Bself) |
| v}" |
| |
| C:\Users\jenkinsSA\AppData\Local\Jenkins |
| \.jenkins\workspace\usermgmtautomation_m |
| aster\.pyenv-python\Lib\site-packages\se |
| mantic_release\helpers.py:52 in _wrapper |
| |
| 49 ) |
| 50 |
| 51 # Call function |
| > 52 result = func(*args, * |
| 53 |
| 54 # Log result |
| 55 logger.debug("%s -> %s |
| |
| C:\Users\jenkinsSA\AppData\Local\Jenkins |
| \.jenkins\workspace\usermgmtautomation_m |
| aster\.pyenv-python\Lib\site-packages\se |
| mantic_release\hvcs\gitlab.py:118 in |
| edit_release_notes |
| |
| 115 client.auth() |
| 116 log.info("Updating release |
| 117 |
| > 118 client.projects.get(self.o |
| 119 release_id, |
| 120 { |
| 121 "description": rel |
| |
| C:\Users\jenkinsSA\AppData\Local\Jenkins |
| \.jenkins\workspace\usermgmtautomation_m |
| aster\.pyenv-python\Lib\site-packages\gi |
| tlab\exceptions.py:338 in wrapped_f |
| |
| 335 try: |
| 336 return f(*args, ** |
| 337 except GitlabHttpError |
| > 338 raise error(e.erro |
| 339 |
| 340 return cast(__F, wrapped_f |
| 341 |
+------------------------------------------+
GitlabUpdateError: 403: insufficient_scope
Usage: semantic-release version [OPTIONS]
Try 'semantic-release version -h' for help.
Error: 403: insufficient_scope
```
## Specifications
- python-gitlab version: python_gitlab-3.15.0-py3-none-any.whl
- API version you are using (v3/v4):
- Gitlab server version (or gitlab.com): gitlab.com
|
closed
|
2023-11-27T08:12:16Z
|
2024-12-02T01:54:49Z
|
https://github.com/python-gitlab/python-gitlab/issues/2732
|
[
"support"
] |
damnmso
| 3
|
indico/indico
|
sqlalchemy
| 6,602
|
Peer-review module: having a synthetic table of the paper status
|
**Is your feature request related to a problem? Please describe.**
For the IPAC conferences Light Peer Review we use the indico Peer-Review module. We have more than 1000 papers submitted to the conference and more than 100 submitted to light peer review.
In the judging area we get a list of all papers dominated by the "not submitted" papers.
Having a synthetic table giving the number of paper with the number of paper with each status would help.
**Describe the solution you'd like**
Have somewhere in the peer-review module with the number of papers with each status and a link to see all the papers with that status.
**Describe alternatives you've considered**
For IPAC'23 I made a script that downloaded the page https://indico.jacow.org/event/37/papers/judging/ and counted the number of papers with each status.
**Additional context**
This is the list I am referring to. That page would count the number of papers with each "state"value.
<img width="654" alt="copie_ecran 2024-11-06 à 15 20 01" src="https://github.com/user-attachments/assets/ea635556-e0df-438c-8c8d-2a121cff459b">
|
open
|
2024-11-06T14:51:38Z
|
2024-11-06T14:51:38Z
|
https://github.com/indico/indico/issues/6602
|
[
"enhancement"
] |
NicolasDelerueLAL
| 0
|
iterative/dvc
|
machine-learning
| 10,612
|
"appending" dvc.log.lock file type idea for exps
|
I've been using dvc for a while and I understand the architecture and philosophy. I also understand the decisions behind `dvc exp`eriments with the challenge there being trying to align git's linear development notion with experiment's more "parallel" notion.
I think having to go through dvc exp is too "heavy". Now, if you think of using special git refs as "backend" storage for experiments, alternatively you can achieve the same with dvc lock files that append (instead of overwrite) runs. This way you can keep original (simple) dvc behavior of just dealing with lock files while also capturing the same information for parameterized runs (exps).
So, I should be able to "checkout" a particular run from the dvc.log.lock file instead of having to deal with dvc exp (parameterized or not).
|
closed
|
2024-11-05T17:01:04Z
|
2024-11-05T19:47:28Z
|
https://github.com/iterative/dvc/issues/10612
|
[] |
majidaldo
| 0
|
scikit-learn/scikit-learn
|
data-science
| 30,744
|
Unexpected <class 'AttributeError'>. 'LinearRegression' object has no attribute 'positive
|
My team changed to scikit-learn v1.6.1 this week. We had v1.5.1 before. Our code crashes in this exact line with the error "Unexpected <class 'AttributeError'>. 'LinearRegression' object has no attribute 'positive'".
We cannot deploy in production because of this. I am desperate enough to come here to ask for help. I do not understand why it would complain that the attribute does not exist given that we were using v1.5.1 before and the attribute has existed for 4 years now. My only guess is if we are loading a very old pickled model that does not have the attribute, so it crashes here. Unfortunately I cannot share any pieces of code as it is proprietary.
_Originally posted by @ItsIronOxide in https://github.com/scikit-learn/scikit-learn/pull/30187#discussion_r1937427235_
|
open
|
2025-01-31T15:08:46Z
|
2025-02-04T06:58:02Z
|
https://github.com/scikit-learn/scikit-learn/issues/30744
|
[
"Needs Reproducible Code"
] |
ItsIronOxide
| 2
|
replicate/cog
|
tensorflow
| 2,035
|
replecate.helpers.fileoutput
|
replecate.helpers.fileoutput
i got this error when try to use fooocus ai from replecate api
but the image is complete processing in the server
|
closed
|
2024-10-30T13:09:22Z
|
2024-11-04T13:28:48Z
|
https://github.com/replicate/cog/issues/2035
|
[] |
uptimeai11062024
| 1
|
InstaPy/InstaPy
|
automation
| 6,826
|
Instagram
|
<!-- Did you know that we have a Discord channel ? Join us: https://discord.gg/FDETsht -->
<!-- Is this a Feature Request ? Please, check out our Wiki first https://github.com/timgrossmann/InstaPy/wiki -->
## Expected Behavior
## Current Behavior
## Possible Solution (optional)
## InstaPy
> **configuration**
|
open
|
2024-09-20T20:04:52Z
|
2024-09-20T20:04:52Z
|
https://github.com/InstaPy/InstaPy/issues/6826
|
[] |
R299489
| 0
|
explosion/spaCy
|
machine-learning
| 13,635
|
Still encounter "TypeError: issubclass() arg 1 must be a class" problem with pydantic == 2.9.2 and scapy == 3.7.6
|
### Discussed in https://github.com/explosion/spaCy/discussions/13634
<div type='discussions-op-text'>
<sup>Originally posted by **LuoXiaoxi-cxq** September 26, 2024</sup>
I was running the training part of the [self-attentive-parser] (https://github.com/nikitakit/self-attentive-parser), where I met this error (probably caused by version problems of `spacy` and `pydantic`):
```
Traceback (most recent call last):
File "D:\postgraduate\research\parsing\self-attentive-parser\src\main.py", line 11, in <module>
from benepar import char_lstm
File "D:\postgraduate\research\parsing\self-attentive-parser\src\benepar\__init__.py", line 20, in <module>
from .integrations.spacy_plugin import BeneparComponent, NonConstituentException
File "D:\postgraduate\research\parsing\self-attentive-parser\src\benepar\integrations\spacy_plugin.py", line 5, in <module>
from .spacy_extensions import ConstituentData, NonConstituentException
File "D:\postgraduate\research\parsing\self-attentive-parser\src\benepar\integrations\spacy_extensions.py", line 177, in <module>
install_spacy_extensions()
File "D:\postgraduate\research\parsing\self-attentive-parser\src\benepar\integrations\spacy_extensions.py", line 153, in install_spacy_extensions
from spacy.tokens import Doc, Span, Token
File "D:\anaconda\lib\site-packages\spacy\__init__.py", line 14, in <module>
from . import pipeline # noqa: F401
File "D:\anaconda\lib\site-packages\spacy\pipeline\__init__.py", line 1, in <module>
from .attributeruler import AttributeRuler
File "D:\anaconda\lib\site-packages\spacy\pipeline\attributeruler.py", line 6, in <module>
from .pipe import Pipe
File "spacy\pipeline\pipe.pyx", line 8, in init spacy.pipeline.pipe
File "D:\anaconda\lib\site-packages\spacy\training\__init__.py", line 11, in <module>
from .callbacks import create_copy_from_base_model # noqa: F401
File "D:\anaconda\lib\site-packages\spacy\training\callbacks.py", line 3, in <module>
from ..language import Language
File "D:\anaconda\lib\site-packages\spacy\language.py", line 25, in <module>
from .training.initialize import init_vocab, init_tok2vec
File "D:\anaconda\lib\site-packages\spacy\training\initialize.py", line 14, in <module>
from .pretrain import get_tok2vec_ref
File "D:\anaconda\lib\site-packages\spacy\training\pretrain.py", line 16, in <module>
from ..schemas import ConfigSchemaPretrain
File "D:\anaconda\lib\site-packages\spacy\schemas.py", line 216, in <module>
class TokenPattern(BaseModel):
File "pydantic\main.py", line 299, in pydantic.main.ModelMetaclass.__new__
print("Loaded {:,} test examples.".format(len(test_treebank)))
File "pydantic\fields.py", line 411, in pydantic.fields.ModelField.infer
File "pydantic\fields.py", line 342, in pydantic.fields.ModelField.__init__
File "pydantic\fields.py", line 451, in pydantic.fields.ModelField.prepare
File "pydantic\fields.py", line 545, in pydantic.fields.ModelField._type_analysis
File "pydantic\fields.py", line 550, in pydantic.fields.ModelField._type_analysis
File "D:\anaconda\lib\typing.py", line 852, in __subclasscheck__
return issubclass(cls, self.__origin__)
TypeError: issubclass() arg 1 must be a class
```
[This issue](https://github.com/langchain-ai/langchain/issues/7522) says installing two packages `chromadb` and `pydantic` will work, so I installed them. I ran
```
python -m pip install -U pydantic spacy
python -m pip install -U chromadb spacy
```
I am running on Windows 11. Now, my environment is
```
Python == 3.10.14
pydantic == 2.9.2
pydantic-core == 2.23.4
scapy == 3.7.6
typing-extensions == 4.12.2
chromadb == 0.5.9
```
However, the problem still exists. According to [this issue](https://github.com/explosion/spaCy/issues/12659), the problem ("TypeError: issubclass() arg 1 must be a class") should only exist for `pydantic` v1.10.7 and earlier related to the recent release of `typing_extensions` v4.6.0. My versions are higher, but this error isn't solved.</div>
|
open
|
2024-09-26T08:24:35Z
|
2024-09-26T08:25:47Z
|
https://github.com/explosion/spaCy/issues/13635
|
[] |
LuoXiaoxi-cxq
| 0
|
unionai-oss/pandera
|
pandas
| 1,893
|
DataFrameSchema is not hashable
|
**Describe the bug**
`DataFrameSchema` is not hashable, and therefore cannot be used by e.g. functools.cache
- [X] I have checked that this issue has not already been reported.
- [X] I have confirmed this bug exists on the latest version of pandera.
- [ ] (optional) I have confirmed this bug exists on the main branch of pandera.
#### Code Sample, a copy-pastable example
```python
import pandera as pa
schema = pa.DataFrameSchema({
"column1": pa.Column(int)
})
hash(schema)
```
#### Expected behavior
Given that `DataFrameSchema` have good `__str__` and `__repr__`s I'd have thought it would be straightforward to make them hashable, and therefore usable as dictionary keys and in turn cache'able.
As a local hack, I added the following monkey-patch
```python
DataFrameSchema.__hash__ = lambda self: hash(repr(self))
```
which doesn't seem too terrible?
#### Desktop (please complete the following information):
- OS: WSL Ubuntu 24.04 LTS on Windows 11
- Browser: N/A
- Version: 0.22.1
#### Screenshots
N/A
#### Additional context
N/A
|
open
|
2025-01-07T14:49:25Z
|
2025-01-07T14:49:25Z
|
https://github.com/unionai-oss/pandera/issues/1893
|
[
"bug"
] |
sparkiegeek
| 0
|
CorentinJ/Real-Time-Voice-Cloning
|
pytorch
| 443
|
How can i pass a root directory for datasets ?
|
datasets_root: None
enc_models_dir: encoder\saved_models
syn_models_dir: synthesizer\saved_models
voc_models_dir: vocoder\saved_models
low_mem: False
Warning: you did not pass a root directory for datasets as argument.
When i open the toolbox,i can't add any projections,pls help
|
closed
|
2020-07-24T01:21:32Z
|
2020-07-24T06:29:20Z
|
https://github.com/CorentinJ/Real-Time-Voice-Cloning/issues/443
|
[] |
FulvioZozix
| 3
|
dmlc/gluon-nlp
|
numpy
| 835
|
Export script for transformer
|
Currently the transformer decoder reads the shape of the input to decide if it runs the inference mode or evaluation mode. We can probably add an export script to properly export a HybridBlock. @sxjscience @szhengac
For masks generation, that can be handled by the `contrib.arange_like` or `arange(max_len)` + `slice_like` already.
|
open
|
2019-07-17T22:30:42Z
|
2019-07-18T12:37:42Z
|
https://github.com/dmlc/gluon-nlp/issues/835
|
[
"enhancement"
] |
eric-haibin-lin
| 1
|
LAION-AI/Open-Assistant
|
machine-learning
| 2,705
|
Add reward model scoring during SFT evaluation
|
Currently we still need to manually run [sampling_score.py](https://github.com/LAION-AI/Open-Assistant/blob/main/model/model_eval/sampling_score.py) on our [sampling reports](https://open-assistant.github.io/oasst-model-eval/?f=https%3A%2F%2Fraw.githubusercontent.com%2FOpen-Assistant%2Foasst-model-eval%2Fmain%2Fsampling_reports%2Foasst-sft%2F2023-04-17_OpenAssistant_oasst-sft-7e2-llama-30b_sampling_noprefix2.json%0A%0A) after training. In order to simplify the evaluation process and to get a score from our RM earlier during training this evaluation should be integrated directly in the SFT training process as an additional evaluation metric that is reported to wandb. Currently only classic evaluation is done which computes the loss and accuracy scores of an evaluation set.
For RL we trained several reward models which can be found on our Huggingface page: 1.4B pythia-based: [oasst-rm-2.1-pythia-1.4b-epoch-2.5](https://huggingface.co/OpenAssistant/oasst-rm-2.1-pythia-1.4b-epoch-2.5), 6.9B pythia-based: [OpenAssistant/oasst-rm-2-pythia-6.9b-epoch-1](https://huggingface.co/OpenAssistant/oasst-rm-2-pythia-6.9b-epoch-1). On the HF page a short snippet is shown how to load the RM and how to compute a reward score for samples.
Several things should be considered:
1. Memory usage: The reward model does not need to stay on the GPU the whole time, it could either be kept completely in CPU memory or only be loaded into the GPU for the eval step.
2. The RM model uses the tokenizer configuration of Pythia while we also train models with other tokenizers that have different EOS token representations (e.g. pythia `<|endoftext|>` vs. llama `</s>`). For a meaningful evaluation the special tokens of the trained model replaced to the pythia ones before tokenization. In general it is not possible to directly forward the token-ids but a token to text-conversion of the trained model need to happen followed by again a tokenization of this text to feed it into the RM.
If you are to the codebase looking at [trainer_sft.py](https://github.com/LAION-AI/Open-Assistant/blob/main/model/model_training/trainer_sft.py) and checking how to add a custom step into the [HF Trainer](https://huggingface.co/docs/transformers/main_classes/trainer) derived class would probably be a good first step.
|
open
|
2023-04-18T09:20:39Z
|
2023-06-06T01:51:52Z
|
https://github.com/LAION-AI/Open-Assistant/issues/2705
|
[
"ml"
] |
andreaskoepf
| 4
|
ploomber/ploomber
|
jupyter
| 487
|
Improve warnings management when running ploomber build
|
Sometimes, third-party libraries emit warnings; when ploomber runs a pipeline, it will capture the warnings and show them at the end.
- [ ] see if we can do something to improve the information displayed to help the user find what library displayed the warning
- [ ] add a way to suppress the warnings. it's possible to not capture them by switching the executor, so maybe add a section in the docs and print a link it if dag executes with warnings
|
closed
|
2022-01-17T00:11:45Z
|
2023-06-21T21:40:46Z
|
https://github.com/ploomber/ploomber/issues/487
|
[] |
edublancas
| 1
|
ultralytics/ultralytics
|
computer-vision
| 19,827
|
How to Freeze Detection Head Layers in YOLOv8m-segment and Train Only Segmentation Head?
|
### Search before asking
- [x] I have searched the Ultralytics YOLO [issues](https://github.com/ultralytics/ultralytics/issues) and [discussions](https://github.com/orgs/ultralytics/discussions) and found no similar questions.
### Question
Question:
Hi all,
I'm working with yolov8m-seg.pt and want to freeze the detection head layers (bounding box/class prediction) while training only the segmentation head (mask prediction). The goal is to fine-tune the segmentation capability without updating the detection part. Has anyone done this before?
I’m thinking of freezing layers by setting requires_grad = False for detection-related params, but I’m unsure how to precisely identify them in the head (e.g., model.22). Here’s my tentative code—can someone confirm if this approach works or suggest a better way?
### Additional
`from ultralytics import YOLO
# Load model
model = YOLO("yolov8m-seg.pt")
# Freeze detection head layers (guessing these are related to 'detect')
for name, param in model.model.named_parameters():
if "detect" in name.lower(): # Is this the right way to target detection head?
param.requires_grad = False
# Train only segmentation head
model.train(data="path/to/data.yaml", epochs=50, imgsz=640)`
Questions:
Does detect correctly target the detection head, or should I use a different identifier (e.g., specific layer indices)?
Will this setup ensure the segmentation head (e.g., mask coefficients/Proto) still trains properly?
Any pitfalls to watch out for?
Thanks for any insights!
|
closed
|
2025-03-23T04:50:37Z
|
2025-03-24T00:49:05Z
|
https://github.com/ultralytics/ultralytics/issues/19827
|
[
"question",
"segment"
] |
Wang-taoshuo
| 3
|
SkalskiP/courses
|
computer-vision
| 31
|
Please add a license?
|
Would love to add/reuse this repo in other projects, but without a license I am unable to use it. Please consider adding a license?
|
open
|
2023-05-25T14:11:50Z
|
2023-05-29T15:16:16Z
|
https://github.com/SkalskiP/courses/issues/31
|
[
"question"
] |
tosaddler
| 2
|
proplot-dev/proplot
|
data-visualization
| 231
|
Box plot and Violin plot do not show a column, if the column contain NaN values
|
<!-- Thanks for helping us make proplot a better package! If this is a bug report, please use the template provided below. If this is a feature request, you can delete the template text (just try to be descriptive with your request). -->
### Description
Box plot and Violin plots do not show the columns which contain NaN values.
### Steps to reproduce
```python
import proplot as plot
import numpy as np
import pandas as pd
# Generate sample data
N = 500
state = np.random.RandomState(51423)
data = state.normal(size=(N, 5)) + 2 * (state.rand(N, 5) - 0.5) * np.arange(5)
data = pd.DataFrame(
data,
columns=pd.Index(['a', 'b', 'c', 'd', 'e'], name='xlabel')
)
#Adding a NaN value to column 'e'
new_row = {'a':2, 'b':2, 'c':2, 'd':2, 'e':np.nan}
data = data.append(new_row, ignore_index=True)
# Generate figure
fig, axs = plot.subplots(ncols=2, axwidth=2.5)
axs.format(grid=False, suptitle='Boxes and violins demo')
# Box plots
ax = axs[0]
obj1 = ax.boxplot(
data, lw=0.7, marker='x', fillcolor='gray5',
medianlw=1, mediancolor='k'
)
ax.format(title='Box plots', titleloc='uc')
# Violin plots
ax = axs[1]
obj2 = ax.violinplot(
data, lw=0.7, fillcolor='gray7',
points=500, bw_method=0.3, means=True
)
ax.format(title='Violin plots', titleloc='uc')
```
**Expected behavior**: [What you expected to happen]
**Actual behavior**: [What actually happened]

### Equivalent steps in matplotlib
Please make sure this bug is related to a specific proplot feature. If you're not sure, try to replicate it with the [native matplotlib API](https://matplotlib.org/3.1.1/api/index.html). Matplotlib bugs belong on the [matplotlib github page](https://github.com/matplotlib/matplotlib).
```python
# your code here, if applicable
```
### Proplot version
6.4.0
|
closed
|
2020-10-29T12:13:20Z
|
2021-06-29T20:36:14Z
|
https://github.com/proplot-dev/proplot/issues/231
|
[
"support"
] |
pratiman-91
| 1
|
keras-team/keras
|
deep-learning
| 21,073
|
keras ordinal loss
|
Hi,
could anyone please give me an insight about what the ordinal loss defined in keras represents?
https://www.tensorflow.org/ranking/api_docs/python/tfr/keras/losses/OrdinalLoss
I'm not able to give an interpretation to that definition, as I don't understand for instance what is I_yi > j.
In the documentation there is no citation of any artiecle.
However, I'm looking for an objective fuction for image classification with ordinal categories, a sort of image ranking loss.
Thanks
|
open
|
2025-03-20T11:36:57Z
|
2025-03-20T13:57:50Z
|
https://github.com/keras-team/keras/issues/21073
|
[
"type:support"
] |
LucaSCostanzo
| 0
|
gee-community/geemap
|
jupyter
| 1,192
|
Errro Import geemap in Colab
|
Hello,
I am trying to run this notebook https://colab.research.google.com/drive/134G8UXjNiQYMbOfUgXg40Nsn6bIEorWV#scrollTo=Uz3eJMcPmREH and it generates the following error:
AttributeError: module 'tornado.ioloop' has no attribute '_Selectable'
Thank you very much in advance
```
|
closed
|
2022-08-09T13:16:37Z
|
2022-08-09T19:11:30Z
|
https://github.com/gee-community/geemap/issues/1192
|
[
"bug"
] |
josemanuelgis
| 3
|
yihong0618/running_page
|
data-visualization
| 427
|
KEEP 导出数据不包含轨迹图
|
在APP上面看得到轨迹图的,但是用脚本同步到running page的时候。地图就显示 no map data for this run.
REF LINK:
Sync task: https://github.com/Kilerd/running_page/actions/runs/5324115043/jobs/9642943711
Commit data: https://github.com/Kilerd/running_page/commit/67634935339ba192a18d74ea96647ec7fd5d2c6e
website: https://kilerd.github.io/running_page/
|
closed
|
2023-06-20T15:03:21Z
|
2023-11-01T14:20:35Z
|
https://github.com/yihong0618/running_page/issues/427
|
[] |
Kilerd
| 9
|
keras-rl/keras-rl
|
tensorflow
| 290
|
All policies select_action() func return value of 0 or 1
|
Hi all,
I'm trying to setup a reinforcement learning project using Gym & kears_rl.
**_Description:_**
Given a numbers in a range (100, 200), I want the agent to alert me when a number is close to the limits, lets say between 0%-10% and 90%-100% of the quantiles.
**_Environment info:_**
The environment reward:
quantile [0, 0.1] -> reward = 1
quantile [0.1, 0.9] -> reward = -1
quantile [0.9, 1] -> reward = 1
The agent need to learn the 10% & 90% limits values.
Here is the `observation_space `& `action_space `properties in my environment.py:
```
low = np.array([100])
high = np.array([200])
self.action_space = spaces.Box(low=low, high=high, dtype=np.intt32)
self.observation_space = spaces.Box(low=low, high=high, dtype=np.int32)
```
**_main.py info:_**
```
if __name__ == '__main__':
env = Env(max_steps=100)
nb_actions = env.action_space.shape[0] # equal to 100
model = Sequential()
model.add(Flatten(input_shape=(1,) + env.observation_space.shape))
model.add(Dense(8, activation='relu'))
model.add(Dense(8, activation='relu'))
model.add(Dense(2, activation='softmax'))
memory = SequentialMemory(limit=50000, window_length=1)
policy = BoltzmannQPolicy()
# Create DQN agent
dqn = DQNAgent(model=env.model,
memory=memory,
policy=policy,
nb_actions=nb_actions,
nb_steps_warmup=10,
target_model_update=1e-2)
# Compile the DQN agent
dqn.compile(Adam(lr=1e-3), metrics=['mae'])
# Okay, now it's time to learn something!
dqn.fit(env, nb_steps=50000, visualize=False, verbose=1)
```
**_Questions\issues:_**
1. On the fit function (rl/core.py:169), the `action `I get equal to zero. it should be between [100, 200]. Why is that? I expect the action to be within the action_space but I see that all policies return a value of 0 or 1. How I suppose to use the value in the env.step() func?
It sounds simple, but I just can't get my head around this one... :(
Any help is much appreciated.
Thanks.
|
closed
|
2019-01-30T11:43:44Z
|
2019-05-07T17:31:50Z
|
https://github.com/keras-rl/keras-rl/issues/290
|
[
"wontfix"
] |
yaniv11386
| 1
|
Anjok07/ultimatevocalremovergui
|
pytorch
| 1,439
|
使用uvr5产生报错
|
下面是报错文本
Last Error Received:
Process: VR Architecture
If this error persists, please contact the developers with the error details.
Raw Error Details:
MemoryError: "Unable to allocate 1.43 GiB for an array with shape (2, 769, 125046) and data type complex64"
Traceback Error: "
File "UVR.py", line 6638, in process_start
File "separate.py", line 1055, in seperate
File "separate.py", line 1196, in inference_vr
File "separate.py", line 1173, in postprocess
"
Error Time Stamp [2024-07-01 18:46:21]
Full Application Settings:
vr_model: 3_HP-Vocal-UVR
aggression_setting: 5
window_size: 512
mdx_segment_size: 256
batch_size: Default
crop_size: 256
is_tta: False
is_output_image: False
is_post_process: False
is_high_end_process: False
post_process_threshold: 0.2
vr_voc_inst_secondary_model: No Model Selected
vr_other_secondary_model: No Model Selected
vr_bass_secondary_model: No Model Selected
vr_drums_secondary_model: No Model Selected
vr_is_secondary_model_activate: False
vr_voc_inst_secondary_model_scale: 0.9
vr_other_secondary_model_scale: 0.7
vr_bass_secondary_model_scale: 0.5
vr_drums_secondary_model_scale: 0.5
demucs_model: Choose Model
segment: Default
overlap: 0.25
overlap_mdx: Default
overlap_mdx23: 8
shifts: 2
chunks_demucs: Auto
margin_demucs: 44100
is_chunk_demucs: False
is_chunk_mdxnet: False
is_primary_stem_only_Demucs: False
is_secondary_stem_only_Demucs: False
is_split_mode: True
is_demucs_combine_stems: True
is_mdx23_combine_stems: True
demucs_voc_inst_secondary_model: No Model Selected
demucs_other_secondary_model: No Model Selected
demucs_bass_secondary_model: No Model Selected
demucs_drums_secondary_model: No Model Selected
demucs_is_secondary_model_activate: False
demucs_voc_inst_secondary_model_scale: 0.9
demucs_other_secondary_model_scale: 0.7
demucs_bass_secondary_model_scale: 0.5
demucs_drums_secondary_model_scale: 0.5
demucs_pre_proc_model: No Model Selected
is_demucs_pre_proc_model_activate: False
is_demucs_pre_proc_model_inst_mix: False
mdx_net_model: UVR-MDX-NET Inst HQ 3
chunks: Auto
margin: 44100
compensate: Auto
denoise_option: None
is_match_frequency_pitch: True
phase_option: Automatic
phase_shifts: None
is_save_align: False
is_match_silence: True
is_spec_match: False
is_mdx_c_seg_def: False
is_invert_spec: False
is_deverb_vocals: False
deverb_vocal_opt: Main Vocals Only
voc_split_save_opt: Lead Only
is_mixer_mode: False
mdx_batch_size: Default
mdx_voc_inst_secondary_model: No Model Selected
mdx_other_secondary_model: No Model Selected
mdx_bass_secondary_model: No Model Selected
mdx_drums_secondary_model: No Model Selected
mdx_is_secondary_model_activate: False
mdx_voc_inst_secondary_model_scale: 0.9
mdx_other_secondary_model_scale: 0.7
mdx_bass_secondary_model_scale: 0.5
mdx_drums_secondary_model_scale: 0.5
is_save_all_outputs_ensemble: True
is_append_ensemble_name: False
chosen_audio_tool: Manual Ensemble
choose_algorithm: Min Spec
time_stretch_rate: 2.0
pitch_rate: 2.0
is_time_correction: True
is_gpu_conversion: True
is_primary_stem_only: True
is_secondary_stem_only: False
is_testing_audio: False
is_auto_update_model_params: True
is_add_model_name: False
is_accept_any_input: False
is_task_complete: False
is_normalization: False
is_use_opencl: False
is_wav_ensemble: False
is_create_model_folder: False
mp3_bit_set: 320k
semitone_shift: 0
save_format: WAV
wav_type_set: 32-bit Float
device_set: NVIDIA GeForce RTX 3060 Laptop GPU:0
help_hints_var: True
set_vocal_splitter: No Model Selected
is_set_vocal_splitter: False
is_save_inst_set_vocal_splitter: False
model_sample_mode: False
model_sample_mode_duration: 30
demucs_stems: All Stems
mdx_stems: All Stems
|
open
|
2024-07-01T10:55:13Z
|
2024-07-01T10:56:07Z
|
https://github.com/Anjok07/ultimatevocalremovergui/issues/1439
|
[] |
FanBo555
| 1
|
tatsu-lab/stanford_alpaca
|
deep-learning
| 312
|
openai version
|
May I know which version of openai is used for training? I wonder if which version provides openai_object since the latest one does not support it.
Thank you!
|
closed
|
2024-02-29T09:05:54Z
|
2024-03-02T09:39:32Z
|
https://github.com/tatsu-lab/stanford_alpaca/issues/312
|
[] |
cswangxiaowei
| 1
|
CorentinJ/Real-Time-Voice-Cloning
|
pytorch
| 780
|
pip3 install -r requirements.txt not working
|
when i try to run pip3 install -r requirements.txt it gives me an error saying Command "python setup.py egg_info" failed with error code 1 in /tmp/pip-install-z3vkjilm/PyQt5/
|
closed
|
2021-06-21T02:35:44Z
|
2021-06-21T02:57:34Z
|
https://github.com/CorentinJ/Real-Time-Voice-Cloning/issues/780
|
[] |
smithblaze911
| 0
|
plotly/dash-bio
|
dash
| 28
|
create common user for all dash gallery apps
|
as mentioned in : https://github.com/plotly/streambed/issues/11788#issuecomment-436746312
- [x] see with Chelsea if this user already exists
- [x] create `developers`/`dataliberation` user on dds
- [x] dash-gallery exceeded the 15 person license, checkup with Hamza on that, as new user cannot create apps
- [x] delete /dash-bio app from sham's account (if not there's url conflict..)
- [x] add dash-bio path to common user
- [ ] setup environment requirements for dash-bio app so that Chitra's app works: https://github.com/plotly/streambed/issues/11803
- [x] redeploy master with `developers` as user
- [x] edit pr template with new steps for deployment, now everyone will be responsible for redeploy once they merge to master
|
closed
|
2018-11-11T17:01:23Z
|
2018-11-30T00:52:29Z
|
https://github.com/plotly/dash-bio/issues/28
|
[] |
VeraZab
| 1
|
home-assistant/core
|
python
| 140,591
|
BUG: utility_meter.reset action does not populate recommendation list
|
### The problem
when using utility_meter.reset action in the UI it does not recommend utility meters you have but if you manually enter one by its entity name it does function correctly.
this does not happen with utility_meter.calibrate which works as expected


Spoke with @frenck about this on discord and it looks as if its an old backwards compatibility issue from 3 years ago.
### What version of Home Assistant Core has the issue?
2025.3.2
### What was the last working version of Home Assistant Core?
_No response_
### What type of installation are you running?
Home Assistant OS
### Integration causing the issue
Utility Meter
### Link to integration documentation on our website
https://www.home-assistant.io/integrations/utility_meter/
### Diagnostics information
_No response_
### Example YAML snippet
```yaml
```
### Anything in the logs that might be useful for us?
```txt
```
### Additional information
_No response_
|
open
|
2025-03-14T11:26:31Z
|
2025-03-14T12:19:09Z
|
https://github.com/home-assistant/core/issues/140591
|
[
"integration: utility_meter"
] |
MichaelMKKelly
| 4
|
pytorch/pytorch
|
machine-learning
| 149,828
|
bump XNNPACK dependency to fix GCC 14 build on aarch64-linux
|
### 🐛 Describe the bug
bundled version of XNNPACK cannot be built on aarch64-linux with GCC14 because of this issue https://github.com/google/XNNPACK/issues/7726
the issue has been fixed in XNNPACK in the meanwhile: https://github.com/google/XNNPACK/commit/3bc2a32a44db62434248197bceefa37f4f05153e
suggestion: bump the XNNPACK dependency in `third_party` to newer commit which contains the fix
### Versions
2.6.0
cc @malfet @seemethere @snadampal @milpuz01 @aditew01 @nikhil-arm @fadara01
|
open
|
2025-03-23T15:16:07Z
|
2025-03-24T03:51:04Z
|
https://github.com/pytorch/pytorch/issues/149828
|
[
"module: build",
"triaged",
"actionable",
"module: xnnpack",
"module: arm"
] |
prusnak
| 1
|
Evil0ctal/Douyin_TikTok_Download_API
|
fastapi
| 306
|
[BUG] 抖音获取数据时失败
|
***发生错误的平台?***
如:抖音
***发生错误的端点?***
如:API-V1/API-V2
***提交的输入值?***
如:短视频链接
***是否有再次尝试?***
如:是,发生错误后X时间后错误依旧存在。
***你有查看本项目的自述文件或接口文档吗?***
如:有,并且很确定该问题是程序导致的。
*******报错提示如下***********
ValueError: 获取抖音视频数据出错了: 0, message='Attempt to decode JSON with unexpected mimetype: text/plain; charset=utf-8', url=URL('https://www.douyin.com/aweme/v1/web/aweme/detail/?device_platform=webapp&aid=6383&channel=channel_pc_web&aweme_id=7289724683019111714&pc_client_type=1&version_code=190500&version_name=19.5.0&cookie_enabled=true&screen_width=1344&screen_height=756&browser_language=zh-CN&browser_platform=Win32&browser_name=Firefox&browser_version=118.0&browser_online=true&engine_name=Gecko&engine_version=109.0&os_name=Windows&os_version=10&cpu_core_num=16&device_memory=&platform=PC&webid=7284189800734082615&msToken=B1N9FM825TkvFbayDsDvZxM8r5suLrsfQbC93TciS0O9Iii8iJpAPd__FM2rpLUJi5xtMencSXLeNn8xmOS9q7bP0CUsrt9oVTL08YXLPRzZm0dHKLc9PGRlyEk=&X-Bogus=DFSzswSLaJ0ANnEftYQ3ot9WcBnF')
|
closed
|
2023-10-23T11:27:16Z
|
2024-02-07T03:43:41Z
|
https://github.com/Evil0ctal/Douyin_TikTok_Download_API/issues/306
|
[
"BUG"
] |
jiupinjiandingshi
| 1
|
ckan/ckan
|
api
| 8,092
|
readthedocs sphinx build failures
|
## CKAN version
master
## Describe the bug
infinite loop in build, looks like no tags are returned from `git log`?
### Steps to reproduce
check sphinx logs
### Expected behavior
build docs on rtd working
### Additional details
```python-traceback
Traceback (most recent call last):
File "/home/docs/checkouts/readthedocs.org/user_builds/ckan/envs/latest/lib/python3.10/site-packages/sphinx/config.py", line 358, in eval_config_file
exec(code, namespace) # NoQA: S102
File "/home/docs/checkouts/readthedocs.org/user_builds/ckan/checkouts/latest/doc/conf.py", line 388, in <module>
current_release_tag_value = get_current_release_tag()
File "/home/docs/checkouts/readthedocs.org/user_builds/ckan/checkouts/latest/doc/conf.py", line 211, in get_current_release_tag
return get_latest_release_tag()
File "/home/docs/checkouts/readthedocs.org/user_builds/ckan/checkouts/latest/doc/conf.py", line 228, in get_latest_release_tag
return get_latest_release_version()
File "/home/docs/checkouts/readthedocs.org/user_builds/ckan/checkouts/latest/doc/conf.py", line 237, in get_latest_release_version
version = get_latest_release_tag()[len('ckan-'):]
File "/home/docs/checkouts/readthedocs.org/user_builds/ckan/checkouts/latest/doc/conf.py", line 228, in get_latest_release_tag
return get_latest_release_version()
File "/home/docs/checkouts/readthedocs.org/user_builds/ckan/checkouts/latest/doc/conf.py", line 237, in get_latest_release_version
version = get_latest_release_tag()[len('ckan-'):]
…
```
|
closed
|
2024-02-28T20:29:59Z
|
2024-03-07T09:31:31Z
|
https://github.com/ckan/ckan/issues/8092
|
[] |
wardi
| 0
|
sinaptik-ai/pandas-ai
|
data-visualization
| 1,669
|
Addition of LLM base models
|
### 🚀 The feature
Instead of calling a LLM via API, I want the library to be capable of leveraging base models(Llama, Deepseek etc.) installed in the local machine.
### Motivation, pitch
Hi! I was trying out the library but found myself running out of tokens pretty quickly. I believe that adding an option to add the base models can be really effective for users who want to leverage their computational resources for the task and build their applications.
### Alternatives
_No response_
### Additional context
_No response_
|
closed
|
2025-03-11T15:47:56Z
|
2025-03-14T16:47:12Z
|
https://github.com/sinaptik-ai/pandas-ai/issues/1669
|
[] |
SnehalBhartiya
| 3
|
autogluon/autogluon
|
data-science
| 4,966
|
[BUG] Incorrect learning rate schedule when using multiple GPUs in `MultiModalPredictor`
|
**Bug Report Checklist**
<!-- Please ensure at least one of the following to help the developers troubleshoot the problem: -->
- [x] I provided code that demonstrates a minimal reproducible example. <!-- Ideal, especially via source install -->
- [x] I confirmed bug exists on the latest mainline of AutoGluon via source install. <!-- Preferred -->
- [x] I confirmed bug exists on the latest stable version of AutoGluon. <!-- Unnecessary if prior items are checked -->
**Describe the bug**
<!-- A clear and concise description of what the bug is. -->
Based on [this line](https://github.com/autogluon/autogluon/blob/e1961672e5d34661ab2e4e78dba64a8741cc88f0/multimodal/src/autogluon/multimodal/optimization/lit_module.py#L384) we have that the maximum number of steps for the training of `MultiModalPredictor` is computed as
```
max_steps = (
len(self.trainer.datamodule.train_dataloader())
* self.trainer.max_epochs
// self.trainer.accumulate_grad_batches
)
```
However, `len(self.trainer.datamodule.train_dataloader())` will return the number of steps in one epoch **before considering the number of GPUs used for training**.
**Example**:
1. I have a dataset of 800 samples and a batch size of 10 -> one epoch will take 80 steps.
1. I train on a machine with 4 GPUs -> effective batch size becomes 40 -> one epoch will take 20 steps.
1. I train for 5 epochs -> `max_steps=100`.
1. `MultiModalPredictor` will compute `max_steps=400` and will set the learning rate schedule accordingly.
1. As a consequence, if I choose `warmup_steps=0.1` (so that 10% of total training steps should be devoted to warmup) and `lr_schedule="cosine_decay"` (so that the learning rate should decay up to a value of 0 at the very last training step), I will actually see that **40% of the training steps are devoted to warmup and after that the learning rate decays slowly up to some positive, non-zero value**.
**Expected behavior**
<!-- A clear and concise description of what you expected to happen. -->
In the example above, we would expect to see a linear growth in the learning rate for the first 10% of the training and then a cosine decay up to 0 at the very last step.
**To Reproduce**
<!-- A minimal script to reproduce the issue. Links to Colab notebooks or similar tools are encouraged.
If the code is too long, feel free to put it in a public gist and link it in the issue: https://gist.github.com.
In short, we are going to copy-paste your code to run it and we expect to get the same result as you. -->
Minimal example: [gist](https://gist.github.com/SnoopKilla/cba71dd9f2ce99e4371a51b46b872f57)
**Steps to reproduce**:
1. Run the code on a machine with 4 GPUs available.
2. Check the tensorboard logs for the plotted learning rate schedule and assert that it is not as expected.
**Screenshots / Logs**
<!-- If applicable, add screenshots or logs to help explain your problem. -->
<img width="951" alt="Image" src="https://github.com/user-attachments/assets/eb2c7cca-752b-4595-aa89-c5cb1e6c05b2" />
**Installed Versions**
<!-- Please run the following code snippet: -->
<details>
```
INSTALLED VERSIONS
------------------
date : 2025-03-07
time : 21:52:25.057437
python : 3.11.10.final.0
OS : Linux
OS-release : 4.14.355-275.570.amzn2.x86_64
Version : #1 SMP Sat Nov 30 09:51:35 UTC 2024
machine : x86_64
processor : x86_64
num_cores : 96
cpu_ram_mb : 382905.51953125
cuda version : 12.550.127.05
num_gpus : 4
gpu_ram_mb : [22500, 22245, 22500, 22500]
avail_disk_size_mb : 441560
accelerate : 0.34.2
autogluon : 1.2
autogluon.common : 1.2
autogluon.core : 1.2
autogluon.features : 1.2
autogluon.multimodal : 1.2
autogluon.tabular : 1.2
autogluon.timeseries : 1.2
boto3 : 1.35.63
catboost : 1.2.7
coreforecast : 0.0.12
defusedxml : 0.7.1
einops : 0.8.0
evaluate : 0.4.3
fastai : 2.7.18
fugue : 0.9.1
gluonts : 0.16.0
huggingface-hub : 0.26.2
hyperopt : 0.2.7
imodels : None
jinja2 : 3.1.4
joblib : 1.4.2
jsonschema : 4.21.1
lightgbm : 4.5.0
lightning : 2.4.0
matplotlib : 3.9.2
mlforecast : 0.13.4
networkx : 3.4.2
nlpaug : 1.1.11
nltk : 3.8.1
numpy : 1.26.4
nvidia-ml-py3 : 7.352.0
omegaconf : 2.2.3
onnx : None
onnxruntime : None
onnxruntime-gpu : None
openmim : 0.3.9
optimum : None
optimum-intel : None
orjson : 3.10.12
pandas : 2.2.3
pdf2image : 1.17.0
Pillow : 10.4.0
psutil : 6.1.0
pyarrow : 18.0.0
pytesseract : 0.3.10
pytorch-lightning : 2.4.0
pytorch-metric-learning: 2.3.0
ray : 2.39.0
requests : 2.32.3
scikit-image : 0.24.0
scikit-learn : 1.5.2
scikit-learn-intelex : None
scipy : 1.14.1
seqeval : 1.2.2
skl2onnx : None
spacy : 3.7.5
statsforecast : 1.7.8
tabpfn : None
tensorboard : 2.18.0
text-unidecode : 1.3
timm : 1.0.3
torch : 2.5.1+cu124
torchmetrics : 1.2.1
torchvision : 0.20.1+cu124
tqdm : 4.66.5
transformers : 4.47.1
utilsforecast : 0.2.4
vowpalwabbit : None
xgboost : 2.1.3
None
```
</details>
|
open
|
2025-03-07T21:53:31Z
|
2025-03-07T21:53:31Z
|
https://github.com/autogluon/autogluon/issues/4966
|
[
"bug: unconfirmed",
"Needs Triage"
] |
SnoopKilla
| 0
|
huggingface/text-generation-inference
|
nlp
| 2,670
|
Prefix caching causes 2 different responses from the same HTTP call with seed set depending on what machine calls
|
### System Info
tag:2.3.1 docker image running on nvidia 4090 on top of 20.04 Ubuntu
```
2024-10-18T19:25:04.160854Z INFO text_generation_launcher: Args {
model_id: "Qwen/Qwen2.5-Coder-1.5B",
revision: None,
validation_workers: 2,
sharded: None,
num_shard: None,
quantize: Some(
Fp8,
),
speculate: Some(
6,
),
dtype: None,
trust_remote_code: false,
max_concurrent_requests: 128,
max_best_of: 2,
max_stop_sequences: 4,
max_top_n_tokens: 5,
max_input_tokens: Some(
9000,
),
max_input_length: None,
max_total_tokens: Some(
9999,
),
waiting_served_ratio: 0.3,
max_batch_prefill_tokens: Some(
10000,
),
max_batch_total_tokens: None,
max_waiting_tokens: 20,
max_batch_size: None,
cuda_graphs: None,
hostname: "3f2367249b02",
port: 80,
shard_uds_path: "/tmp/text-generation-server",
master_addr: "localhost",
master_port: 29500,
huggingface_hub_cache: None,
weights_cache_override: None,
disable_custom_kernels: false,
cuda_memory_fraction: 1.0,
rope_scaling: None,
rope_factor: None,
json_output: false,
otlp_endpoint: None,
otlp_service_name: "text-generation-inference.router",
cors_allow_origin: [],
api_key: None,
watermark_gamma: None,
watermark_delta: None,
ngrok: false,
ngrok_authtoken: None,
ngrok_edge: None,
tokenizer_config_path: None,
disable_grammar_support: false,
env: false,
max_client_batch_size: 4,
lora_adapters: None,
usage_stats: On,
}
```
### Information
- [X] Docker
- [ ] The CLI directly
### Tasks
- [X] An officially supported command
- [ ] My own modifications
### Reproduction
Dumped the raw HTTP request from my server that is calling then replicated on my personal machine to the same TGI server and get 2 different responses. I dumped the raw HTTP calls because after validating the payload my only thought was maybe headers but there are no headers included aside from Content-Type and Content-Length. Using fasthttp in golang to make the call. The current example isn't the best since the responses are close. Normally I get garbage from the server call and quality from the local machine call. I tried rolling back to v2.2.0 to exclude prefix caching in case that was the problem but the qwen model is not supported. Is it possible to disable prefix caching to test?
Server
```
POST /generate HTTP/1.1
Host: <REDACTED>:8080
Content-Type: application/json
Content-Length: 789
{"inputs":"\u003c|file_sep|\u003ebot/tts_handler.py\n\u003c|fim_prefix|\u003eimport io\nimport logging\nfrom elevenlabs import generate, Voice, VoiceSettings, set_api_key\nfrom config import Config\n\nlogger= logging.getLogger(__name__)\n\nclass TTSHandler:\n def __init(self, config: Config):\n self.config = config\n self.voice = Voice(config.voice_id)\n self.voice_settings = VoiceSettings(config.voice_settings)\n \u003c|fim_suffix|\u003e\n \n\n\n\u003c|fim_middle|\u003e","parameters":{"do_sample":false,"max_new_tokens":1000,"return_full_text":false,"stop":["\u003c|file_sep|\u003e","\u003c|repo_name|\u003e","\u003c|fim_prefix|\u003e","\n"],"seed":69420,"temperature":0.3,"top_k":50,"top_p":0.8,"watermark":false,"details":true},"stream":false}
```
Response
```
{"generated_text":"set_api_key(config.elevenlabs_api_key1\n","details":{"finish_reason":"stop_sequence","generated_tokens":12,"seed":69420,"prefill":[],"tokens":[{"id":746,"text":"set","logprob":0.0,"special":false},{"id":11697,"text":"_api","logprob":0.0,"special":false},{"id":3097,"text":"_key","logprob":0.0,"special":false},{"id":8754,"text":"(config","logprob":0.0,"special":false},{"id":1734,"text":".e","logprob":0.0,"special":false},{"id":273,"text":"le","logprob":0.0,"special":false},{"id":1037,"text":"ven","logprob":0.0,"special":false},{"id":70271,"text":"labs","logprob":0.0,"special":false},{"id":11697,"text":"_api","logprob":0.0,"special":false},{"id":3097,"text":"_key","logprob":0.0,"special":false},{"id":16,"text":"1","logprob":0.0,"special":false},{"id":198,"text":"\n","logprob":0.0,"special":false}]}}
```
Local
```
POST /generate HTTP/1.1
Host: <REDACTED>:8080
Content-Type: application/json
Content-Length: 789
{"inputs":"\u003c|file_sep|\u003ebot/tts_handler.py\n\u003c|fim_prefix|\u003eimport io\nimport logging\nfrom elevenlabs import generate, Voice, VoiceSettings, set_api_key\nfrom config import Config\n\nlogger= logging.getLogger(__name__)\n\nclass TTSHandler:\n def __init(self, config: Config):\n self.config = config\n self.voice = Voice(config.voice_id)\n self.voice_settings = VoiceSettings(config.voice_settings)\n \u003c|fim_suffix|\u003e\n \n\n\n\u003c|fim_middle|\u003e","parameters":{"do_sample":false,"max_new_tokens":1000,"return_full_text":false,"stop":["\u003c|file_sep|\u003e","\u003c|repo_name|\u003e","\u003c|fim_prefix|\u003e","\n"],"seed":69420,"temperature":0.3,"top_k":50,"top_p":0.8,"watermark":false,"details":true},"stream":false}
```
Response
```
{"generated_text":"set_api_key(config.elevenlabs_api_key(config123456789012\\\n","details":{"finish_reason":"stop_sequence","generated_tokens":25,"seed":69420,"prefill":[],"tokens":[{"id":746,"text":"set","logprob":0.0,"special":false},{"id":11697,"text":"_api","logprob":0.0,"special":false},{"id":3097,"text":"_key","logprob":0.0,"special":false},{"id":8754,"text":"(config","logprob":0.0,"special":false},{"id":1734,"text":".e","logprob":0.0,"special":false},{"id":273,"text":"le","logprob":0.0,"special":false},{"id":1037,"text":"ven","logprob":0.0,"special":false},{"id":70271,"text":"labs","logprob":0.0,"special":false},{"id":11697,"text":"_api","logprob":0.0,"special":false},{"id":3097,"text":"_key","logprob":0.0,"special":false},{"id":8754,"text":"(config","logprob":0.0,"special":false},{"id":16,"text":"1","logprob":0.0,"special":false},{"id":17,"text":"2","logprob":0.0,"special":false},{"id":18,"text":"3","logprob":0.0,"special":false},{"id":19,"text":"4","logprob":0.0,"special":false},{"id":20,"text":"5","
logprob":0.0,"special":false},{"id":21,"text":"6","logprob":0.0,"special":false},{"id":22,"text":"7","logprob":0.0,"special":false},{"id":23,"text":"8","logprob":0.0,"special":false},{"id":24,"text":"9","logprob":0.0,"special":false},{"id":15,"text":"0","logprob":0.0,"special":false},{"id":16,"text":"1","logprob":-0.3125,"special":false},{"id":17,"text":"2","logprob":0.0,"special":false},{"id":59,"text":"\\","logprob":-2.078125,"special":false},{"id":198,"text":"\n","logprob":0.0,"special":false}]}}
```
### Expected behavior
Same response with the same call to the same tgi server regardless of the macine
|
open
|
2024-10-18T21:53:57Z
|
2024-10-23T20:05:06Z
|
https://github.com/huggingface/text-generation-inference/issues/2670
|
[] |
sam-ulrich1
| 5
|
onnx/onnx
|
pytorch
| 6,561
|
No Adapter From Version $17 for GroupNormalization
|
# Ask a Question
### Question
Im trying to convert my onnx model (with GroupNormalization op) from domain version 11 to 21 by using
` converted_model = version_converter.convert_version(model, 21)`
and got error
`line 37, in convert_version
converted_model_str = C.convert_version(model_str, target_version)
RuntimeError: /github/workspace/onnx/version_converter/BaseConverter.h:73: adapter_lookup: Assertion `false` failed: No Adapter From Version $17 for GroupNormalization
`
### Further information
and my onnx version info etc as following
`
>>> import onnx
>>> onnx.__version__
'1.16.0'
>>> onnx.defs.onnx_opset_version()
21
>>>
`
and GN supporting info on onnx official doc is like following

how can i fix this "No adapter" problem? i also tried upgrade onnx==1.17.0, and got the same issue.
|
open
|
2024-11-29T10:44:41Z
|
2024-11-29T10:44:41Z
|
https://github.com/onnx/onnx/issues/6561
|
[
"question"
] |
yzhou0919
| 0
|
healthchecks/healthchecks
|
django
| 535
|
ServiceNow
|
### Discussed in https://github.com/healthchecks/healthchecks/discussions/532
<div type='discussions-op-text'>
<sup>Originally posted by **yellowdigital** June 16, 2021</sup>
Hello, We have a requirement to add in integration to ServiceNow platform.
Initially we had this working using the existing "webhook" integration and posting in a well formed JSON body string to the webhook. This was working well but was not using strong authentication, only basic Auth in the header.
We now need to extend this functionality to allow for an Oauth2 token request phase, then using bearer auth with the returned token, to post the data into the webhook URL.
Would anybody be interested in developing an new integration to handle this scenario? We would be prepared to contribute towards the cost of building this and getting it approved into the public repository healthchecks.</div>
|
open
|
2021-06-22T11:45:38Z
|
2023-07-14T17:53:14Z
|
https://github.com/healthchecks/healthchecks/issues/535
|
[
"new integration"
] |
yellowdigital
| 0
|
deeppavlov/DeepPavlov
|
nlp
| 1,567
|
👩💻📞 DeepPavlov Community Call #16
|
Dear DeepPavlov community,
We are excited to announce that we are back with our Community Calls in English. The next one will be on May 26! Don’t miss the chance to meet with our team and learn more about building Knowledge Graph-infused AI assistants with DeepPavlov Dream.
Daniel Kornev, CPO of DeepPavlov, will talk about DeepPavlov Dream, the first open-source academia-built multiskill AI assistant platform that uses KGs in its skills and in the shared memory of the system. You will find out how the team has integrated KGs into the AI Assistant platform, and how you can leverage it in building your own KG-infused AI assistants today.
As always, we are waiting for your suggestions and hope to see them on Calls!
**DeepPavlov Community Call #16, English Edition (May 26th, 2022)
We’ll hold it on May 26th, 2022 at 16:00 UTC.**
> Add to your calendar:
> https://bit.ly/DPMonthlyCall
We welcome you to join us at our DeepPavlov Community Call #16 to let us know what you think about the latest changes and tell us how DeepPavlov helps you in your projects!
> Agenda for DeepPavlov Community Call #16:
>
> 16:00 –16:10 | Greeting
>
> 16:10 –16:30 | About DeepPavlov Dream
>
> 16:30 –17:00 | DeepPavlov Dream: building knowledge graph-based AI assistants
>
> 17:00 –17:30 | Q&A with DeepPavlov Engineering Team
In case you’ve missed the last one, we’ve uploaded a record — [see the playlist](https://bit.ly/DPCommunityCall13_Video). Check it out!
### **DeepPavlov Library Feedback Survey**
We want to hear your thoughts. You can fill in this form to let us know how you use DeepPavlov Library, what you want us to add or improve!
We are anxious to hear your thoughts!
https://bit.ly/DPLibrarySurvey
### ****Interested?****
Please let us know and leave a comment with any topics or questions you’d like to hear about!
We can’t promise to cover everything but we’ll do our best later this month or in a future call.
After calling in or watching, please do fill in the survey to let us know if you have any feedback on the format or content: https://bit.ly/dpcallsurvey
See you!
The [DeepPavlov](https://deeppavlov.ai/) team
|
closed
|
2022-05-26T12:43:25Z
|
2022-06-16T11:41:29Z
|
https://github.com/deeppavlov/DeepPavlov/issues/1567
|
[
"discussion"
] |
PolinaMrdv
| 0
|
gevent/gevent
|
asyncio
| 1,902
|
PY311: unknown type name ‘CFrame’
|
This error occurred post python-greenlet/greenlet#306
```
building 'gevent._gevent_c_greenlet_primitives' extension
gcc -pthread -Wsign-compare -DNDEBUG -g -fwrapv -O3 -Wall -fPIC -I/opt/hostedtoolcache/Python/3.11.0-rc.1/x64/include/python3.11 -I/opt/hostedtoolcache/Python/3.11.0-rc.1/x64/include/python3.11 -I/tmp/pip-install-vmkvai20/gevent_1002667104154261abd2e40bb0aa2cf6/deps -Isrc/gevent -Isrc/gevent/libev -Isrc/gevent/resolver -I. -I/home/runner/work/geventmp/geventmp/target/venv/build/cpython-3.11.0.candidate.1/include -I/opt/hostedtoolcache/Python/3.11.0-rc.1/x64/include/python3.11 -c src/gevent/_greenlet_primitives.c -o build/temp.linux-x86_64-cpython-311/src/gevent/_greenlet_primitives.o
In file included from src/gevent/_greenlet_primitives.c:1050:
/tmp/pip-install-vmkvai20/gevent_1002667104154261abd2e40bb0aa2cf6/deps/greenlet/greenlet.h:42:5: error: unknown type name ‘CFrame’
42 | CFrame* cframe;
| ^~~~~~
error: command '/usr/bin/gcc' failed with exit code 1
[end of output]
```
https://github.com/karellen/geventmp/runs/8051746537?check_suite_focus=true#step:4:708
|
closed
|
2022-08-27T17:02:08Z
|
2022-10-14T15:05:03Z
|
https://github.com/gevent/gevent/issues/1902
|
[] |
arcivanov
| 0
|
littlecodersh/ItChat
|
api
| 925
|
很好用
|
itchat真的不错
|
closed
|
2020-07-20T02:42:51Z
|
2023-11-16T12:35:45Z
|
https://github.com/littlecodersh/ItChat/issues/925
|
[] |
2905683882
| 2
|
keras-team/keras
|
deep-learning
| 20,576
|
How do I specify a ragged tensor as an input object in a sequential model (Keras v3)
|
I am following the [object detection tutorial with yolov8 and kerascv](https://keras.io/examples/vision/yolov8/)
However, instead of using yolo for object detection, I would like to use a custom sequential model.
My roadblock is at the point of defining the input layer. I would like it to take in a ragged tensor.
According to the [keras v2 docs](https://keras.io/2.16/api/layers/core_layers/input/), there is an option to specify `ragged=True` and define the input layer via `inputs = tf.keras.Input(shape = [] , dtype = tf.int64, ragged = True)`
However in [keras v3](https://keras.io/api/layers/core_layers/input/), the `ragged` option has been removed.
So how do I proceed with defining an input layer that takes in a ragged tensor?
|
closed
|
2024-12-02T06:42:20Z
|
2025-01-31T02:00:17Z
|
https://github.com/keras-team/keras/issues/20576
|
[
"stat:awaiting response from contributor",
"stale",
"type:Bug"
] |
fninsiima
| 4
|
mitmproxy/mitmproxy
|
python
| 6,886
|
mitmdump crashes on dns requests in reverse proxy mode
|
#### Problem Description
I ran a simple mitmdump instance in reverse proxy mode and when I send DNS requests it crashes and stops listening and proccessing packets. I get `Python interoperability task shutting down. Task failed: UDP recv() failed` error in the logs.
This only happens for some domains, for example it doesn't happen for github.com but it happens for google.com. It looks like DNS response retransmission packet is causing the crash.
Also tried mitmproxy and mitmweb, they were no different, tried version 9 and it too crashed. This issue is not present in linux.
#### Steps to reproduce the behavior:
1. mitmdump --mode reverse:dns://1.1.1.1@53
2. nslookup google.com 127.0.0.1
#### System Information
Mitmproxy: 10.3.0 binary
Python: 3.12.3
OpenSSL: OpenSSL 3.2.1 30 Jan 2024
Platform: Windows-10-10.0.19045-SP0
#### Logs
this is the log without debug:
```
>mitmdump --mode reverse:dns://1.1.1.1@53
[02:31:08.275] reverse proxy to dns://1.1.1.1 listening at *:53.
[02:31:16.025][127.0.0.1:51093] client connect
[02:31:16.027][127.0.0.1:51093] server connect 1.1.1.1:53
127.0.0.1:51093: DNS QUERY (PTR) 1.0.0.127.in-addr.arpa
<< NXDOMAIN
[02:31:16.157][127.0.0.1:51095] client connect
[02:31:16.158][127.0.0.1:51095] server connect 1.1.1.1:53
127.0.0.1:51095: DNS QUERY (A) google.com.lan
<< NXDOMAIN
[02:31:16.287][127.0.0.1:51097] client connect
[02:31:16.288][127.0.0.1:51097] server connect 1.1.1.1:53
127.0.0.1:51097: DNS QUERY (AAAA) google.com.lan
<< NXDOMAIN
[02:31:16.415][127.0.0.1:51099] client connect
[02:31:16.417][127.0.0.1:51099] server connect 1.1.1.1:53
127.0.0.1:51099: DNS QUERY (A) google.com
<< 216.239.38.120
[02:31:16.454][127.0.0.1:51101] client connect
[02:31:16.456][127.0.0.1:51101] server connect 1.1.1.1:53
127.0.0.1:51101: DNS QUERY (AAAA) google.com
<< 2001:4860:4802:32::78
127.0.0.1:51099: DNS QUERY (A) google.com
<< 142.250.186.46
[02:31:16.527] Task failed: UDP recv() failed
disabled backtrace
[02:31:16.527][127.0.0.1:51097] server disconnect 1.1.1.1:53
[02:31:16.530][127.0.0.1:51097] client disconnect
[02:31:16.530][127.0.0.1:51093] server disconnect 1.1.1.1:53
[02:31:16.531][127.0.0.1:51099] server disconnect 1.1.1.1:53
[02:31:16.532][127.0.0.1:51093] client disconnect
[02:31:16.532][127.0.0.1:51099] client disconnect
[02:31:16.533][127.0.0.1:51095] server disconnect 1.1.1.1:53
[02:31:16.533][127.0.0.1:51101] server disconnect 1.1.1.1:53
[02:31:16.534][127.0.0.1:51095] client disconnect
[02:31:16.534][127.0.0.1:51101] client disconnect
```
and with debug:
```
>mitmdump --mode reverse:dns://8.8.8.8@53 --set termlog_verbosity=debug --set console_eventlog_verbosity=debug --set proxy_debug=true
[02:33:53.054] Initializing UDP server ...
[02:33:53.055] UDP server listening on 0.0.0.0:53 ...
[02:33:53.056] UDP server successfully initialized.
[02:33:53.071] Initializing UDP server ...
[02:33:53.072] UDP server listening on [::]:53 ...
[02:33:53.073] UDP server successfully initialized.
[02:33:53.073] reverse proxy to dns://8.8.8.8 listening at *:53.
[02:33:56.896][127.0.0.1:64743] client connect
[02:33:56.896][127.0.0.1:64743] >> Start({})
[02:33:56.897][127.0.0.1:64743] >> Start({})
[02:33:56.898][127.0.0.1:64743] >> DataReceived(client, b'\x00\x01\x01\x00\x00\x01\x00\x00\x00\x00\x00\x00\x011\x010\x010\x03127\x07in-addr\x04arpa\x00\x00\x0c\x00\x01')
[02:33:56.899][127.0.0.1:64743] >> DataReceived(client, b'\x00\x01\x01\x00\x00\x01\x00\x00\x00\x00\x00\x00\x011\x010\x010\x03127\x07in-addr\x04arpa\x00\x00\x0c\x00\x01')
[02:33:56.900][127.0.0.1:64743] << NextLayerHook(data=NextLayer:None)
[02:33:56.900][127.0.0.1:64743] << NextLayerHook(data=NextLayer:None)
[02:33:56.901][127.0.0.1:64743] >> Reply(NextLayerHook(data=NextLayer:DNSLayer(state: start)), None)
[02:33:56.901][127.0.0.1:64743] >> Reply(NextLayerHook(data=NextLayer:DNSLayer(state: start)), None)
[02:33:56.901][127.0.0.1:64743] [nextlayer] DNSLayer(state: start)
[02:33:56.902][127.0.0.1:64743] >> Start({})
[02:33:56.903][127.0.0.1:64743] >> DataReceived(client, b'\x00\x01\x01\x00\x00\x01\x00\x00\x00\x00\x00\x00\x011\x010\x010\x03127\x07in-addr\x04arpa\x00\x00\x0c\x00\x01')
[02:33:56.904][127.0.0.1:64743] << DnsRequestHook(flow=<DNSFlow
request=Message(timestamp=1717196636.9047368, id=1, query=True, op_code=0, authoritative_answer=False, truncation=False, recursion_desired=True, recursion_available=False, reserved=0, response_code=0, questions=[Question(name='1.0.0.127.in-addr.arpa', type=12, class_=1)], answers=[], authorities=[], additionals=[])
response=None
>)
[02:33:56.905][127.0.0.1:64743] << DnsRequestHook(flow=<DNSFlow
[02:33:56.905][127.0.0.1:64743] << DnsRequestHook(flow=<DNSFlow
[02:33:56.906][127.0.0.1:64743] >> Reply(DnsRequestHook(flow=<DNSFlow
request=Message(timestamp=1717196636.9047368, id=1, query=True, op_code=0, authoritative_answer=False, truncation=False, recursion_desired=True, recursion_available=False, reserved=0, response_code=0, questions=[Question(name='1.0.0.127.in-addr.arpa', type=12, class_=1)], answers=[], authorities=[], additionals=[])
response=None
>), None)
[02:33:56.906][127.0.0.1:64743] >> Reply(DnsRequestHook(flow=<DNSFlow
[02:33:56.907][127.0.0.1:64743] >> Reply(DnsRequestHook(flow=<DNSFlow
[02:33:56.907][127.0.0.1:64743] << OpenConnection({'connection': Server({'id': '…5eb969', 'address': ('8.8.8.8', 53), 'transport_protocol': 'udp'})})
[02:33:56.907][127.0.0.1:64743] << OpenConnection({'connection': Server({'id': '…5eb969', 'address': ('8.8.8.8', 53), 'transport_protocol': 'udp'})})
[02:33:56.908][127.0.0.1:64743] << OpenConnection({'connection': Server({'id': '…5eb969', 'address': ('8.8.8.8', 53), 'transport_protocol': 'udp'})})
[02:33:56.910][127.0.0.1:64743] server connect 8.8.8.8:53
[02:33:56.914][127.0.0.1:64743] >> Reply(OpenConnection({'connection': Server({'id': '…5eb969', 'address': ('8.8.8.8', 53), 'peername': ('8.8.8.8', 53), 'sockname': ('192.168.0.17', 64744), 'state': <ConnectionState.OPEN: 3>, 'transport_protocol': 'udp', 'timestamp_start': 1717196636.908739})}), None)
[02:33:56.914][127.0.0.1:64743] >> Reply(OpenConnection({'connection': Server({'id': '…5eb969', 'address': ('8.8.8.8', 53), 'peername': ('8.8.8.8', 53), 'sockname': ('192.168.0.17', 64744), 'state': <ConnectionState.OPEN: 3>, 'transport_protocol': 'udp', 'timestamp_start': 1717196636.908…
[02:33:56.915][127.0.0.1:64743] << SendData(server, b'\x00\x01\x01\x00\x00\x01\x00\x00\x00\x00\x00\x00\x011\x010\x010\x03127\x07in-addr\x04arpa\x00\x00\x0c\x00\x01')
[02:33:56.915][127.0.0.1:64743] << SendData(server, b'\x00\x01\x01\x00\x00\x01\x00\x00\x00\x00\x00\x00\x011\x010\x010\x03127\x07in-addr\x04arpa\x00\x00\x0c\x00\x01')
[02:33:56.991][127.0.0.1:64743] >> DataReceived(server, b'\x00\x01\x81\x83\x00\x01\x00\x00\x00\x01\x00\x00\x011\x010\x010\x03127\x07in-addr\x04arpa\x00\x00\x0c\x00\x01\xc0\x16\x00\x06\x00\x01\x00\x00\x07&\x008\x01b\x0fin-addr-servers\xc0\x1e\x05nstld\x04iana\x03org\x00x\x86\xb5L\x00\x00\x07\x08\x00\x00\x03\x84\x00\t:\x80\x00\x00\x0e\x10')
[02:33:56.992][127.0.0.1:64743] >> DataReceived(server, b'\x00\x01\x81\x83\x00\x01\x00\x00\x00\x01\x00\x00\x011\x010\x010\x03127\x07in-addr\x04arpa\x00\x00\x0c\x00\x01\xc0\x16\x00\x06\x00\x01\x00\x00\x07&\x008\x01b\x0fin-addr-servers\xc0\x1e\x05nstld\x04iana\x03org\x00x\x86\xb5L\x00\x00\…
[02:33:56.993][127.0.0.1:64743] << DnsResponseHook(flow=<DNSFlow
request=Message(timestamp=1717196636.9047368, id=1, query=True, op_code=0, authoritative_answer=False, truncation=False, recursion_desired=True, recursion_available=False, reserved=0, response_code=0, questions=[Question(name='1.0.0.127.in-addr.arpa', type=12, class_=1)], answers=[], authorities=[], additionals=[])
response=Message(timestamp=1717196636.9930813, id=1, query=False, op_code=0, authoritative_answer=False, truncation=False, recursion_desired=True, recursion…
[02:33:56.996][127.0.0.1:64743] << DnsResponseHook(flow=<DNSFlow
127.0.0.1:64743: DNS QUERY (PTR) 1.0.0.127.in-addr.arpa
<< NXDOMAIN
[02:33:56.997][127.0.0.1:64743] >> Reply(DnsResponseHook(flow=<DNSFlow
request=Message(timestamp=1717196636.9047368, id=1, query=True, op_code=0, authoritative_answer=False, truncation=False, recursion_desired=True, recursion_available=False, reserved=0, response_code=0, questions=[Question(name='1.0.0.127.in-addr.arpa', type=12, class_=1)], answers=[], authorities=[], additionals=[])
response=Message(timestamp=1717196636.9930813, id=1, query=False, op_code=0, authoritative_answer=False, truncation=False, recursion_desired=True, rec…
[02:33:56.998][127.0.0.1:64743] >> Reply(DnsResponseHook(flow=<DNSFlow
[02:33:56.998][127.0.0.1:64743] << SendData(client, b'\x00\x01\x81\x83\x00\x01\x00\x00\x00\x01\x00\x00\x011\x010\x010\x03127\x07in-addr\x04arpa\x00\x00\x0c\x00\x01\x07in-addr\x04arpa\x00\x00\x06\x00\x01\x00\x00\x07&\x008\x01b\x0fin-addr-servers\xc0\x1e\x05nstld\x04iana\x03org\x00x\x86\xb5L\x00\x00\x07\x08\x00\x00\x03\x84\x00\t:\x80\x00\x00\x0e\x10')
[02:33:56.999][127.0.0.1:64743] << SendData(client, b'\x00\x01\x81\x83\x00\x01\x00\x00\x00\x01\x00\x00\x011\x010\x010\x03127\x07in-addr\x04arpa\x00\x00\x0c\x00\x01\x07in-addr\x04arpa\x00\x00\x06\x00\x01\x00\x00\x07&\x008\x01b\x0fin-addr-servers\xc0\x1e\x05nstld\x04iana\x03org\x00x\x86\xb…
[02:33:57.001][127.0.0.1:64745] client connect
[02:33:57.001][127.0.0.1:64745] >> Start({})
[02:33:57.002][127.0.0.1:64745] >> Start({})
[02:33:57.003][127.0.0.1:64745] >> DataReceived(client, b'\x00\x02\x01\x00\x00\x01\x00\x00\x00\x00\x00\x00\x06google\x03com\x03lan\x00\x00\x01\x00\x01')
[02:33:57.003][127.0.0.1:64745] >> DataReceived(client, b'\x00\x02\x01\x00\x00\x01\x00\x00\x00\x00\x00\x00\x06google\x03com\x03lan\x00\x00\x01\x00\x01')
[02:33:57.003][127.0.0.1:64745] << NextLayerHook(data=NextLayer:None)
[02:33:57.004][127.0.0.1:64745] << NextLayerHook(data=NextLayer:None)
[02:33:57.004][127.0.0.1:64745] >> Reply(NextLayerHook(data=NextLayer:DNSLayer(state: start)), None)
[02:33:57.004][127.0.0.1:64745] >> Reply(NextLayerHook(data=NextLayer:DNSLayer(state: start)), None)
[02:33:57.005][127.0.0.1:64745] [nextlayer] DNSLayer(state: start)
[02:33:57.008][127.0.0.1:64745] >> Start({})
[02:33:57.008][127.0.0.1:64745] >> DataReceived(client, b'\x00\x02\x01\x00\x00\x01\x00\x00\x00\x00\x00\x00\x06google\x03com\x03lan\x00\x00\x01\x00\x01')
[02:33:57.009][127.0.0.1:64745] << DnsRequestHook(flow=<DNSFlow
request=Message(timestamp=1717196637.0096836, id=2, query=True, op_code=0, authoritative_answer=False, truncation=False, recursion_desired=True, recursion_available=False, reserved=0, response_code=0, questions=[Question(name='google.com.lan', type=1, class_=1)], answers=[], authorities=[], additionals=[])
response=None
>)
[02:33:57.010][127.0.0.1:64745] << DnsRequestHook(flow=<DNSFlow
[02:33:57.010][127.0.0.1:64745] << DnsRequestHook(flow=<DNSFlow
[02:33:57.011][127.0.0.1:64745] >> Reply(DnsRequestHook(flow=<DNSFlow
request=Message(timestamp=1717196637.0096836, id=2, query=True, op_code=0, authoritative_answer=False, truncation=False, recursion_desired=True, recursion_available=False, reserved=0, response_code=0, questions=[Question(name='google.com.lan', type=1, class_=1)], answers=[], authorities=[], additionals=[])
response=None
>), None)
[02:33:57.011][127.0.0.1:64745] >> Reply(DnsRequestHook(flow=<DNSFlow
[02:33:57.012][127.0.0.1:64745] >> Reply(DnsRequestHook(flow=<DNSFlow
[02:33:57.013][127.0.0.1:64745] << OpenConnection({'connection': Server({'id': '…599044', 'address': ('8.8.8.8', 53), 'transport_protocol': 'udp'})})
[02:33:57.013][127.0.0.1:64745] << OpenConnection({'connection': Server({'id': '…599044', 'address': ('8.8.8.8', 53), 'transport_protocol': 'udp'})})
[02:33:57.013][127.0.0.1:64745] << OpenConnection({'connection': Server({'id': '…599044', 'address': ('8.8.8.8', 53), 'transport_protocol': 'udp'})})
[02:33:57.015][127.0.0.1:64745] server connect 8.8.8.8:53
[02:33:57.016][127.0.0.1:64745] >> Reply(OpenConnection({'connection': Server({'id': '…599044', 'address': ('8.8.8.8', 53), 'peername': ('8.8.8.8', 53), 'sockname': ('192.168.0.17', 64746), 'state': <ConnectionState.OPEN: 3>, 'transport_protocol': 'udp', 'timestamp_start': 1717196637.0141444})}), None)
[02:33:57.016][127.0.0.1:64745] >> Reply(OpenConnection({'connection': Server({'id': '…599044', 'address': ('8.8.8.8', 53), 'peername': ('8.8.8.8', 53), 'sockname': ('192.168.0.17', 64746), 'state': <ConnectionState.OPEN: 3>, 'transport_protocol': 'udp', 'timestamp_start': 1717196637.014…
[02:33:57.016][127.0.0.1:64745] << SendData(server, b'\x00\x02\x01\x00\x00\x01\x00\x00\x00\x00\x00\x00\x06google\x03com\x03lan\x00\x00\x01\x00\x01')
[02:33:57.016][127.0.0.1:64745] << SendData(server, b'\x00\x02\x01\x00\x00\x01\x00\x00\x00\x00\x00\x00\x06google\x03com\x03lan\x00\x00\x01\x00\x01')
[02:33:57.224][127.0.0.1:64745] >> DataReceived(server, b'\x00\x02\x81\x83\x00\x01\x00\x00\x00\x01\x00\x00\x06google\x03com\x03lan\x00\x00\x01\x00\x01\x00\x00\x06\x00\x01\x00\x01Qk\x00@\x01a\x0croot-servers\x03net\x00\x05nstld\x0cverisign-grs\x03com\x00x\xa4\x99n\x00\x00\x07\x08\x00\x00\x03\x84\x00\t:\x80\x00\x01Q\x80')
[02:33:57.224][127.0.0.1:64745] >> DataReceived(server, b'\x00\x02\x81\x83\x00\x01\x00\x00\x00\x01\x00\x00\x06google\x03com\x03lan\x00\x00\x01\x00\x01\x00\x00\x06\x00\x01\x00\x01Qk\x00@\x01a\x0croot-servers\x03net\x00\x05nstld\x0cverisign-grs\x03com\x00x\xa4\x99n\x00\x00\x07\x08\x00\x00\…
[02:33:57.226][127.0.0.1:64745] << DnsResponseHook(flow=<DNSFlow
request=Message(timestamp=1717196637.0096836, id=2, query=True, op_code=0, authoritative_answer=False, truncation=False, recursion_desired=True, recursion_available=False, reserved=0, response_code=0, questions=[Question(name='google.com.lan', type=1, class_=1)], answers=[], authorities=[], additionals=[])
response=Message(timestamp=1717196637.226156, id=2, query=False, op_code=0, authoritative_answer=False, truncation=False, recursion_desired=True, recursion_available…
[02:33:57.237][127.0.0.1:64745] << DnsResponseHook(flow=<DNSFlow
127.0.0.1:64745: DNS QUERY (A) google.com.lan
<< NXDOMAIN
[02:33:57.239][127.0.0.1:64745] >> Reply(DnsResponseHook(flow=<DNSFlow
request=Message(timestamp=1717196637.0096836, id=2, query=True, op_code=0, authoritative_answer=False, truncation=False, recursion_desired=True, recursion_available=False, reserved=0, response_code=0, questions=[Question(name='google.com.lan', type=1, class_=1)], answers=[], authorities=[], additionals=[])
response=Message(timestamp=1717196637.226156, id=2, query=False, op_code=0, authoritative_answer=False, truncation=False, recursion_desired=True, recursion_ava…
[02:33:57.240][127.0.0.1:64745] >> Reply(DnsResponseHook(flow=<DNSFlow
[02:33:57.241][127.0.0.1:64745] << SendData(client, b'\x00\x02\x81\x83\x00\x01\x00\x00\x00\x01\x00\x00\x06google\x03com\x03lan\x00\x00\x01\x00\x01\x00\x00\x06\x00\x01\x00\x01Qk\x00@\x01a\x0croot-servers\x03net\x00\x05nstld\x0cverisign-grs\x03com\x00x\xa4\x99n\x00\x00\x07\x08\x00\x00\x03\x84\x00\t:\x80\x00\x01Q\x80')
[02:33:57.241][127.0.0.1:64745] << SendData(client, b'\x00\x02\x81\x83\x00\x01\x00\x00\x00\x01\x00\x00\x06google\x03com\x03lan\x00\x00\x01\x00\x01\x00\x00\x06\x00\x01\x00\x01Qk\x00@\x01a\x0croot-servers\x03net\x00\x05nstld\x0cverisign-grs\x03com\x00x\xa4\x99n\x00\x00\x07\x08\x00\x00\x03\…
[02:33:57.242][127.0.0.1:64747] client connect
[02:33:57.243][127.0.0.1:64747] >> Start({})
[02:33:57.243][127.0.0.1:64747] >> Start({})
[02:33:57.244][127.0.0.1:64747] >> DataReceived(client, b'\x00\x03\x01\x00\x00\x01\x00\x00\x00\x00\x00\x00\x06google\x03com\x03lan\x00\x00\x1c\x00\x01')
[02:33:57.244][127.0.0.1:64747] >> DataReceived(client, b'\x00\x03\x01\x00\x00\x01\x00\x00\x00\x00\x00\x00\x06google\x03com\x03lan\x00\x00\x1c\x00\x01')
[02:33:57.244][127.0.0.1:64747] << NextLayerHook(data=NextLayer:None)
[02:33:57.244][127.0.0.1:64747] << NextLayerHook(data=NextLayer:None)
[02:33:57.245][127.0.0.1:64747] >> Reply(NextLayerHook(data=NextLayer:DNSLayer(state: start)), None)
[02:33:57.245][127.0.0.1:64747] >> Reply(NextLayerHook(data=NextLayer:DNSLayer(state: start)), None)
[02:33:57.246][127.0.0.1:64747] [nextlayer] DNSLayer(state: start)
[02:33:57.246][127.0.0.1:64747] >> Start({})
[02:33:57.246][127.0.0.1:64747] >> DataReceived(client, b'\x00\x03\x01\x00\x00\x01\x00\x00\x00\x00\x00\x00\x06google\x03com\x03lan\x00\x00\x1c\x00\x01')
[02:33:57.247][127.0.0.1:64747] << DnsRequestHook(flow=<DNSFlow
request=Message(timestamp=1717196637.2477021, id=3, query=True, op_code=0, authoritative_answer=False, truncation=False, recursion_desired=True, recursion_available=False, reserved=0, response_code=0, questions=[Question(name='google.com.lan', type=28, class_=1)], answers=[], authorities=[], additionals=[])
response=None
>)
[02:33:57.247][127.0.0.1:64747] << DnsRequestHook(flow=<DNSFlow
[02:33:57.248][127.0.0.1:64747] << DnsRequestHook(flow=<DNSFlow
[02:33:57.248][127.0.0.1:64747] >> Reply(DnsRequestHook(flow=<DNSFlow
request=Message(timestamp=1717196637.2477021, id=3, query=True, op_code=0, authoritative_answer=False, truncation=False, recursion_desired=True, recursion_available=False, reserved=0, response_code=0, questions=[Question(name='google.com.lan', type=28, class_=1)], answers=[], authorities=[], additionals=[])
response=None
>), None)
[02:33:57.249][127.0.0.1:64747] >> Reply(DnsRequestHook(flow=<DNSFlow
[02:33:57.249][127.0.0.1:64747] >> Reply(DnsRequestHook(flow=<DNSFlow
[02:33:57.249][127.0.0.1:64747] << OpenConnection({'connection': Server({'id': '…fb45f4', 'address': ('8.8.8.8', 53), 'transport_protocol': 'udp'})})
[02:33:57.250][127.0.0.1:64747] << OpenConnection({'connection': Server({'id': '…fb45f4', 'address': ('8.8.8.8', 53), 'transport_protocol': 'udp'})})
[02:33:57.250][127.0.0.1:64747] << OpenConnection({'connection': Server({'id': '…fb45f4', 'address': ('8.8.8.8', 53), 'transport_protocol': 'udp'})})
[02:33:57.252][127.0.0.1:64747] server connect 8.8.8.8:53
[02:33:57.252][127.0.0.1:64747] >> Reply(OpenConnection({'connection': Server({'id': '…fb45f4', 'address': ('8.8.8.8', 53), 'peername': ('8.8.8.8', 53), 'sockname': ('192.168.0.17', 64748), 'state': <ConnectionState.OPEN: 3>, 'transport_protocol': 'udp', 'timestamp_start': 1717196637.2506948})}), None)
[02:33:57.252][127.0.0.1:64747] >> Reply(OpenConnection({'connection': Server({'id': '…fb45f4', 'address': ('8.8.8.8', 53), 'peername': ('8.8.8.8', 53), 'sockname': ('192.168.0.17', 64748), 'state': <ConnectionState.OPEN: 3>, 'transport_protocol': 'udp', 'timestamp_start': 1717196637.250…
[02:33:57.257][127.0.0.1:64747] << SendData(server, b'\x00\x03\x01\x00\x00\x01\x00\x00\x00\x00\x00\x00\x06google\x03com\x03lan\x00\x00\x1c\x00\x01')
[02:33:57.257][127.0.0.1:64747] << SendData(server, b'\x00\x03\x01\x00\x00\x01\x00\x00\x00\x00\x00\x00\x06google\x03com\x03lan\x00\x00\x1c\x00\x01')
[02:33:57.323][127.0.0.1:64747] >> DataReceived(server, b'\x00\x03\x81\x83\x00\x01\x00\x00\x00\x01\x00\x00\x06google\x03com\x03lan\x00\x00\x1c\x00\x01\x00\x00\x06\x00\x01\x00\x01Q~\x00@\x01a\x0croot-servers\x03net\x00\x05nstld\x0cverisign-grs\x03com\x00x\xa4\x99n\x00\x00\x07\x08\x00\x00\x03\x84\x00\t:\x80\x00\x01Q\x80')
[02:33:57.324][127.0.0.1:64747] >> DataReceived(server, b'\x00\x03\x81\x83\x00\x01\x00\x00\x00\x01\x00\x00\x06google\x03com\x03lan\x00\x00\x1c\x00\x01\x00\x00\x06\x00\x01\x00\x01Q~\x00@\x01a\x0croot-servers\x03net\x00\x05nstld\x0cverisign-grs\x03com\x00x\xa4\x99n\x00\x00\x07\x08\x00\x00\…
[02:33:57.325][127.0.0.1:64747] << DnsResponseHook(flow=<DNSFlow
request=Message(timestamp=1717196637.2477021, id=3, query=True, op_code=0, authoritative_answer=False, truncation=False, recursion_desired=True, recursion_available=False, reserved=0, response_code=0, questions=[Question(name='google.com.lan', type=28, class_=1)], answers=[], authorities=[], additionals=[])
response=Message(timestamp=1717196637.325521, id=3, query=False, op_code=0, authoritative_answer=False, truncation=False, recursion_desired=True, recursion_availabl…
[02:33:57.327][127.0.0.1:64747] << DnsResponseHook(flow=<DNSFlow
127.0.0.1:64747: DNS QUERY (AAAA) google.com.lan
<< NXDOMAIN
[02:33:57.328][127.0.0.1:64747] >> Reply(DnsResponseHook(flow=<DNSFlow
request=Message(timestamp=1717196637.2477021, id=3, query=True, op_code=0, authoritative_answer=False, truncation=False, recursion_desired=True, recursion_available=False, reserved=0, response_code=0, questions=[Question(name='google.com.lan', type=28, class_=1)], answers=[], authorities=[], additionals=[])
response=Message(timestamp=1717196637.325521, id=3, query=False, op_code=0, authoritative_answer=False, truncation=False, recursion_desired=True, recursion_av…
[02:33:57.329][127.0.0.1:64747] >> Reply(DnsResponseHook(flow=<DNSFlow
[02:33:57.329][127.0.0.1:64747] << SendData(client, b'\x00\x03\x81\x83\x00\x01\x00\x00\x00\x01\x00\x00\x06google\x03com\x03lan\x00\x00\x1c\x00\x01\x00\x00\x06\x00\x01\x00\x01Q~\x00@\x01a\x0croot-servers\x03net\x00\x05nstld\x0cverisign-grs\x03com\x00x\xa4\x99n\x00\x00\x07\x08\x00\x00\x03\x84\x00\t:\x80\x00\x01Q\x80')
[02:33:57.330][127.0.0.1:64747] << SendData(client, b'\x00\x03\x81\x83\x00\x01\x00\x00\x00\x01\x00\x00\x06google\x03com\x03lan\x00\x00\x1c\x00\x01\x00\x00\x06\x00\x01\x00\x01Q~\x00@\x01a\x0croot-servers\x03net\x00\x05nstld\x0cverisign-grs\x03com\x00x\xa4\x99n\x00\x00\x07\x08\x00\x00\x03\…
[02:33:57.331][127.0.0.1:64749] client connect
[02:33:57.331][127.0.0.1:64749] >> Start({})
[02:33:57.332][127.0.0.1:64749] >> Start({})
[02:33:57.334][127.0.0.1:64749] >> DataReceived(client, b'\x00\x04\x01\x00\x00\x01\x00\x00\x00\x00\x00\x00\x06google\x03com\x00\x00\x01\x00\x01')
[02:33:57.334][127.0.0.1:64749] >> DataReceived(client, b'\x00\x04\x01\x00\x00\x01\x00\x00\x00\x00\x00\x00\x06google\x03com\x00\x00\x01\x00\x01')
[02:33:57.334][127.0.0.1:64749] << NextLayerHook(data=NextLayer:None)
[02:33:57.335][127.0.0.1:64749] << NextLayerHook(data=NextLayer:None)
[02:33:57.335][127.0.0.1:64749] >> Reply(NextLayerHook(data=NextLayer:DNSLayer(state: start)), None)
[02:33:57.335][127.0.0.1:64749] >> Reply(NextLayerHook(data=NextLayer:DNSLayer(state: start)), None)
[02:33:57.336][127.0.0.1:64749] [nextlayer] DNSLayer(state: start)
[02:33:57.336][127.0.0.1:64749] >> Start({})
[02:33:57.337][127.0.0.1:64749] >> DataReceived(client, b'\x00\x04\x01\x00\x00\x01\x00\x00\x00\x00\x00\x00\x06google\x03com\x00\x00\x01\x00\x01')
[02:33:57.337][127.0.0.1:64749] << DnsRequestHook(flow=<DNSFlow
request=Message(timestamp=1717196637.3370032, id=4, query=True, op_code=0, authoritative_answer=False, truncation=False, recursion_desired=True, recursion_available=False, reserved=0, response_code=0, questions=[Question(name='google.com', type=1, class_=1)], answers=[], authorities=[], additionals=[])
response=None
>)
[02:33:57.338][127.0.0.1:64749] << DnsRequestHook(flow=<DNSFlow
[02:33:57.338][127.0.0.1:64749] << DnsRequestHook(flow=<DNSFlow
[02:33:57.338][127.0.0.1:64749] >> Reply(DnsRequestHook(flow=<DNSFlow
request=Message(timestamp=1717196637.3370032, id=4, query=True, op_code=0, authoritative_answer=False, truncation=False, recursion_desired=True, recursion_available=False, reserved=0, response_code=0, questions=[Question(name='google.com', type=1, class_=1)], answers=[], authorities=[], additionals=[])
response=None
>), None)
[02:33:57.339][127.0.0.1:64749] >> Reply(DnsRequestHook(flow=<DNSFlow
[02:33:57.339][127.0.0.1:64749] >> Reply(DnsRequestHook(flow=<DNSFlow
[02:33:57.339][127.0.0.1:64749] << OpenConnection({'connection': Server({'id': '…918d60', 'address': ('8.8.8.8', 53), 'transport_protocol': 'udp'})})
[02:33:57.340][127.0.0.1:64749] << OpenConnection({'connection': Server({'id': '…918d60', 'address': ('8.8.8.8', 53), 'transport_protocol': 'udp'})})
[02:33:57.340][127.0.0.1:64749] << OpenConnection({'connection': Server({'id': '…918d60', 'address': ('8.8.8.8', 53), 'transport_protocol': 'udp'})})
[02:33:57.342][127.0.0.1:64749] server connect 8.8.8.8:53
[02:33:57.342][127.0.0.1:64749] >> Reply(OpenConnection({'connection': Server({'id': '…918d60', 'address': ('8.8.8.8', 53), 'peername': ('8.8.8.8', 53), 'sockname': ('192.168.0.17', 64750), 'state': <ConnectionState.OPEN: 3>, 'transport_protocol': 'udp', 'timestamp_start': 1717196637.3415804})}), None)
[02:33:57.343][127.0.0.1:64749] >> Reply(OpenConnection({'connection': Server({'id': '…918d60', 'address': ('8.8.8.8', 53), 'peername': ('8.8.8.8', 53), 'sockname': ('192.168.0.17', 64750), 'state': <ConnectionState.OPEN: 3>, 'transport_protocol': 'udp', 'timestamp_start': 1717196637.341…
[02:33:57.343][127.0.0.1:64749] << SendData(server, b'\x00\x04\x01\x00\x00\x01\x00\x00\x00\x00\x00\x00\x06google\x03com\x00\x00\x01\x00\x01')
[02:33:57.344][127.0.0.1:64749] << SendData(server, b'\x00\x04\x01\x00\x00\x01\x00\x00\x00\x00\x00\x00\x06google\x03com\x00\x00\x01\x00\x01')
[02:33:57.376][127.0.0.1:64749] >> DataReceived(server, b'\x00\x04\x81\x80\x00\x01\x00\x01\x00\x00\x00\x00\x06google\x03com\x00\x00\x01\x00\x01\xc0\x0c\x00\x01\x00\x01\x00\x00\x00<\x00\x04\xd8\xef&x')
[02:33:57.377][127.0.0.1:64749] >> DataReceived(server, b'\x00\x04\x81\x80\x00\x01\x00\x01\x00\x00\x00\x00\x06google\x03com\x00\x00\x01\x00\x01\xc0\x0c\x00\x01\x00\x01\x00\x00\x00<\x00\x04\xd8\xef&x')
[02:33:57.379][127.0.0.1:64749] << DnsResponseHook(flow=<DNSFlow
request=Message(timestamp=1717196637.3370032, id=4, query=True, op_code=0, authoritative_answer=False, truncation=False, recursion_desired=True, recursion_available=False, reserved=0, response_code=0, questions=[Question(name='google.com', type=1, class_=1)], answers=[], authorities=[], additionals=[])
response=Message(timestamp=1717196637.378082, id=4, query=False, op_code=0, authoritative_answer=False, truncation=False, recursion_desired=True, recursion_available=Tru…
[02:33:57.380][127.0.0.1:64749] << DnsResponseHook(flow=<DNSFlow
127.0.0.1:64749: DNS QUERY (A) google.com
<< 216.239.38.120
[02:33:57.381][127.0.0.1:64749] >> Reply(DnsResponseHook(flow=<DNSFlow
request=Message(timestamp=1717196637.3370032, id=4, query=True, op_code=0, authoritative_answer=False, truncation=False, recursion_desired=True, recursion_available=False, reserved=0, response_code=0, questions=[Question(name='google.com', type=1, class_=1)], answers=[], authorities=[], additionals=[])
response=Message(timestamp=1717196637.378082, id=4, query=False, op_code=0, authoritative_answer=False, truncation=False, recursion_desired=True, recursion_availab…
[02:33:57.382][127.0.0.1:64749] >> Reply(DnsResponseHook(flow=<DNSFlow
[02:33:57.382][127.0.0.1:64749] << SendData(client, b'\x00\x04\x81\x80\x00\x01\x00\x01\x00\x00\x00\x00\x06google\x03com\x00\x00\x01\x00\x01\x06google\x03com\x00\x00\x01\x00\x01\x00\x00\x00<\x00\x04\xd8\xef&x')
[02:33:57.382][127.0.0.1:64749] << SendData(client, b'\x00\x04\x81\x80\x00\x01\x00\x01\x00\x00\x00\x00\x06google\x03com\x00\x00\x01\x00\x01\x06google\x03com\x00\x00\x01\x00\x01\x00\x00\x00<\x00\x04\xd8\xef&x')
[02:33:57.385][127.0.0.1:64751] client connect
[02:33:57.386][127.0.0.1:64751] >> Start({})
[02:33:57.386][127.0.0.1:64751] >> Start({})
[02:33:57.387][127.0.0.1:64751] >> DataReceived(client, b'\x00\x05\x01\x00\x00\x01\x00\x00\x00\x00\x00\x00\x06google\x03com\x00\x00\x1c\x00\x01')
[02:33:57.387][127.0.0.1:64751] >> DataReceived(client, b'\x00\x05\x01\x00\x00\x01\x00\x00\x00\x00\x00\x00\x06google\x03com\x00\x00\x1c\x00\x01')
[02:33:57.388][127.0.0.1:64751] << NextLayerHook(data=NextLayer:None)
[02:33:57.388][127.0.0.1:64751] << NextLayerHook(data=NextLayer:None)
[02:33:57.389][127.0.0.1:64751] >> Reply(NextLayerHook(data=NextLayer:DNSLayer(state: start)), None)
[02:33:57.389][127.0.0.1:64751] >> Reply(NextLayerHook(data=NextLayer:DNSLayer(state: start)), None)
[02:33:57.389][127.0.0.1:64751] [nextlayer] DNSLayer(state: start)
[02:33:57.390][127.0.0.1:64751] >> Start({})
[02:33:57.390][127.0.0.1:64751] >> DataReceived(client, b'\x00\x05\x01\x00\x00\x01\x00\x00\x00\x00\x00\x00\x06google\x03com\x00\x00\x1c\x00\x01')
[02:33:57.390][127.0.0.1:64751] << DnsRequestHook(flow=<DNSFlow
request=Message(timestamp=1717196637.3909478, id=5, query=True, op_code=0, authoritative_answer=False, truncation=False, recursion_desired=True, recursion_available=False, reserved=0, response_code=0, questions=[Question(name='google.com', type=28, class_=1)], answers=[], authorities=[], additionals=[])
response=None
>)
[02:33:57.391][127.0.0.1:64751] << DnsRequestHook(flow=<DNSFlow
[02:33:57.391][127.0.0.1:64751] << DnsRequestHook(flow=<DNSFlow
[02:33:57.392][127.0.0.1:64751] >> Reply(DnsRequestHook(flow=<DNSFlow
request=Message(timestamp=1717196637.3909478, id=5, query=True, op_code=0, authoritative_answer=False, truncation=False, recursion_desired=True, recursion_available=False, reserved=0, response_code=0, questions=[Question(name='google.com', type=28, class_=1)], answers=[], authorities=[], additionals=[])
response=None
>), None)
[02:33:57.393][127.0.0.1:64751] >> Reply(DnsRequestHook(flow=<DNSFlow
[02:33:57.397][127.0.0.1:64751] >> Reply(DnsRequestHook(flow=<DNSFlow
[02:33:57.397][127.0.0.1:64751] << OpenConnection({'connection': Server({'id': '…ef76b5', 'address': ('8.8.8.8', 53), 'transport_protocol': 'udp'})})
[02:33:57.397][127.0.0.1:64751] << OpenConnection({'connection': Server({'id': '…ef76b5', 'address': ('8.8.8.8', 53), 'transport_protocol': 'udp'})})
[02:33:57.398][127.0.0.1:64751] << OpenConnection({'connection': Server({'id': '…ef76b5', 'address': ('8.8.8.8', 53), 'transport_protocol': 'udp'})})
[02:33:57.400][127.0.0.1:64751] server connect 8.8.8.8:53
[02:33:57.400][127.0.0.1:64751] >> Reply(OpenConnection({'connection': Server({'id': '…ef76b5', 'address': ('8.8.8.8', 53), 'peername': ('8.8.8.8', 53), 'sockname': ('192.168.0.17', 64752), 'state': <ConnectionState.OPEN: 3>, 'transport_protocol': 'udp', 'timestamp_start': 1717196637.3989928})}), None)
[02:33:57.400][127.0.0.1:64751] >> Reply(OpenConnection({'connection': Server({'id': '…ef76b5', 'address': ('8.8.8.8', 53), 'peername': ('8.8.8.8', 53), 'sockname': ('192.168.0.17', 64752), 'state': <ConnectionState.OPEN: 3>, 'transport_protocol': 'udp', 'timestamp_start': 1717196637.398…
[02:33:57.401][127.0.0.1:64751] << SendData(server, b'\x00\x05\x01\x00\x00\x01\x00\x00\x00\x00\x00\x00\x06google\x03com\x00\x00\x1c\x00\x01')
[02:33:57.401][127.0.0.1:64751] << SendData(server, b'\x00\x05\x01\x00\x00\x01\x00\x00\x00\x00\x00\x00\x06google\x03com\x00\x00\x1c\x00\x01')
[02:33:57.414][127.0.0.1:64749] >> DataReceived(server, b'\x00\x04\x81\x80\x00\x01\x00\x01\x00\x00\x00\x00\x06google\x03com\x00\x00\x01\x00\x01\xc0\x0c\x00\x01\x00\x01\x00\x00\x00,\x00\x04\xac\xd9\x11N')
[02:33:57.414][127.0.0.1:64749] >> DataReceived(server, b'\x00\x04\x81\x80\x00\x01\x00\x01\x00\x00\x00\x00\x06google\x03com\x00\x00\x01\x00\x01\xc0\x0c\x00\x01\x00\x01\x00\x00\x00,\x00\x04\xac\xd9\x11N')
[02:33:57.416][127.0.0.1:64749] << DnsResponseHook(flow=<DNSFlow
request=Message(timestamp=1717196637.3370032, id=4, query=True, op_code=0, authoritative_answer=False, truncation=False, recursion_desired=True, recursion_available=False, reserved=0, response_code=0, questions=[Question(name='google.com', type=1, class_=1)], answers=[], authorities=[], additionals=[])
response=Message(timestamp=1717196637.4160051, id=4, query=False, op_code=0, authoritative_answer=False, truncation=False, recursion_desired=True, recursion_available=Tr…
[02:33:57.417][127.0.0.1:64749] << DnsResponseHook(flow=<DNSFlow
127.0.0.1:64749: DNS QUERY (A) google.com
<< 172.217.17.78
[02:33:57.418][127.0.0.1:64749] >> Reply(DnsResponseHook(flow=<DNSFlow
request=Message(timestamp=1717196637.3370032, id=4, query=True, op_code=0, authoritative_answer=False, truncation=False, recursion_desired=True, recursion_available=False, reserved=0, response_code=0, questions=[Question(name='google.com', type=1, class_=1)], answers=[], authorities=[], additionals=[])
response=Message(timestamp=1717196637.4160051, id=4, query=False, op_code=0, authoritative_answer=False, truncation=False, recursion_desired=True, recursion_availa…
[02:33:57.420][127.0.0.1:64749] >> Reply(DnsResponseHook(flow=<DNSFlow
[02:33:57.420][127.0.0.1:64749] << SendData(client, b'\x00\x04\x81\x80\x00\x01\x00\x01\x00\x00\x00\x00\x06google\x03com\x00\x00\x01\x00\x01\x06google\x03com\x00\x00\x01\x00\x01\x00\x00\x00,\x00\x04\xac\xd9\x11N')
[02:33:57.421][127.0.0.1:64749] << SendData(client, b'\x00\x04\x81\x80\x00\x01\x00\x01\x00\x00\x00\x00\x06google\x03com\x00\x00\x01\x00\x01\x06google\x03com\x00\x00\x01\x00\x01\x00\x00\x00,\x00\x04\xac\xd9\x11N')
[02:33:57.422] Python interoperability task shutting down.
[02:33:57.422] Task failed: UDP recv() failed
disabled backtrace
[02:33:57.422][127.0.0.1:64747] >> ConnectionClosed(connection=Client({'id': '…9b2509', 'address': None, 'peername': ('127.0.0.1', 64747), 'sockname': ('0.0.0.0', 53), 'transport_protocol': 'udp', 'timestamp_start': 1717196637.2426941, 'proxy_mode': ProxyMode.parse('reverse:dns://8.8.8.8@53')}))
[02:33:57.423][127.0.0.1:64747] >> ConnectionClosed(connection=Client({'id': '…9b2509', 'address': None, 'peername': ('127.0.0.1', 64747), 'sockname': ('0.0.0.0', 53), 'transport_protocol': 'udp', 'timestamp_start': 1717196637.2426941, 'proxy_mode': ProxyMode.parse('reverse:dns://8.8.8.8…
[02:33:57.424][127.0.0.1:64747] << CloseConnection({'connection': Server({'id': '…fb45f4', 'address': ('8.8.8.8', 53), 'peername': ('8.8.8.8', 53), 'sockname': ('192.168.0.17', 64748), 'state': <ConnectionState.OPEN: 3>, 'transport_protocol': 'udp', 'timestamp_start': 1717196637.2506948})})
[02:33:57.428][127.0.0.1:64747] << CloseConnection({'connection': Server({'id': '…fb45f4', 'address': ('8.8.8.8', 53), 'peername': ('8.8.8.8', 53), 'sockname': ('192.168.0.17', 64748), 'state': <ConnectionState.OPEN: 3>, 'transport_protocol': 'udp', 'timestamp_start': 1717196637.2506948}…
[02:33:57.428][127.0.0.1:64749] >> ConnectionClosed(connection=Client({'id': '…0187a5', 'address': None, 'peername': ('127.0.0.1', 64749), 'sockname': ('0.0.0.0', 53), 'transport_protocol': 'udp', 'timestamp_start': 1717196637.3318255, 'proxy_mode': ProxyMode.parse('reverse:dns://8.8.8.8@53')}))
[02:33:57.429][127.0.0.1:64749] >> ConnectionClosed(connection=Client({'id': '…0187a5', 'address': None, 'peername': ('127.0.0.1', 64749), 'sockname': ('0.0.0.0', 53), 'transport_protocol': 'udp', 'timestamp_start': 1717196637.3318255, 'proxy_mode': ProxyMode.parse('reverse:dns://8.8.8.8…
[02:33:57.430][127.0.0.1:64749] << CloseConnection({'connection': Server({'id': '…918d60', 'address': ('8.8.8.8', 53), 'peername': ('8.8.8.8', 53), 'sockname': ('192.168.0.17', 64750), 'state': <ConnectionState.OPEN: 3>, 'transport_protocol': 'udp', 'timestamp_start': 1717196637.3415804})})
[02:33:57.430][127.0.0.1:64749] << CloseConnection({'connection': Server({'id': '…918d60', 'address': ('8.8.8.8', 53), 'peername': ('8.8.8.8', 53), 'sockname': ('192.168.0.17', 64750), 'state': <ConnectionState.OPEN: 3>, 'transport_protocol': 'udp', 'timestamp_start': 1717196637.3415804}…
[02:33:57.431][127.0.0.1:64747] >> ConnectionClosed(connection=Server({'id': '…fb45f4', 'address': ('8.8.8.8', 53), 'peername': ('8.8.8.8', 53), 'sockname': ('192.168.0.17', 64748), 'transport_protocol': 'udp', 'timestamp_start': 1717196637.2506948}))
[02:33:57.431][127.0.0.1:64747] >> ConnectionClosed(connection=Server({'id': '…fb45f4', 'address': ('8.8.8.8', 53), 'peername': ('8.8.8.8', 53), 'sockname': ('192.168.0.17', 64748), 'transport_protocol': 'udp', 'timestamp_start': 1717196637.2506948}))
[02:33:57.431][127.0.0.1:64747] server disconnect 8.8.8.8:53
[02:33:57.431] UDP client task shutting down.
[02:33:57.432][127.0.0.1:64749] >> ConnectionClosed(connection=Server({'id': '…918d60', 'address': ('8.8.8.8', 53), 'peername': ('8.8.8.8', 53), 'sockname': ('192.168.0.17', 64750), 'transport_protocol': 'udp', 'timestamp_start': 1717196637.3415804}))
[02:33:57.432][127.0.0.1:64749] >> ConnectionClosed(connection=Server({'id': '…918d60', 'address': ('8.8.8.8', 53), 'peername': ('8.8.8.8', 53), 'sockname': ('192.168.0.17', 64750), 'transport_protocol': 'udp', 'timestamp_start': 1717196637.3415804}))
[02:33:57.433][127.0.0.1:64749] server disconnect 8.8.8.8:53
[02:33:57.433] UDP client task shutting down.
[02:33:57.433][127.0.0.1:64743] >> ConnectionClosed(connection=Client({'id': '…bf686d', 'address': None, 'peername': ('127.0.0.1', 64743), 'sockname': ('0.0.0.0', 53), 'transport_protocol': 'udp', 'timestamp_start': 1717196636.8954246, 'proxy_mode': ProxyMode.parse('reverse:dns://8.8.8.8@53')}))
[02:33:57.434][127.0.0.1:64743] >> ConnectionClosed(connection=Client({'id': '…bf686d', 'address': None, 'peername': ('127.0.0.1', 64743), 'sockname': ('0.0.0.0', 53), 'transport_protocol': 'udp', 'timestamp_start': 1717196636.8954246, 'proxy_mode': ProxyMode.parse('reverse:dns://8.8.8.8…
[02:33:57.434][127.0.0.1:64743] << CloseConnection({'connection': Server({'id': '…5eb969', 'address': ('8.8.8.8', 53), 'peername': ('8.8.8.8', 53), 'sockname': ('192.168.0.17', 64744), 'state': <ConnectionState.OPEN: 3>, 'transport_protocol': 'udp', 'timestamp_start': 1717196636.908739})})
[02:33:57.435][127.0.0.1:64743] << CloseConnection({'connection': Server({'id': '…5eb969', 'address': ('8.8.8.8', 53), 'peername': ('8.8.8.8', 53), 'sockname': ('192.168.0.17', 64744), 'state': <ConnectionState.OPEN: 3>, 'transport_protocol': 'udp', 'timestamp_start': 1717196636.908739})…
[02:33:57.435][127.0.0.1:64745] >> ConnectionClosed(connection=Client({'id': '…01f92f', 'address': None, 'peername': ('127.0.0.1', 64745), 'sockname': ('0.0.0.0', 53), 'transport_protocol': 'udp', 'timestamp_start': 1717196637.0013015, 'proxy_mode': ProxyMode.parse('reverse:dns://8.8.8.8@53')}))
[02:33:57.435][127.0.0.1:64745] >> ConnectionClosed(connection=Client({'id': '…01f92f', 'address': None, 'peername': ('127.0.0.1', 64745), 'sockname': ('0.0.0.0', 53), 'transport_protocol': 'udp', 'timestamp_start': 1717196637.0013015, 'proxy_mode': ProxyMode.parse('reverse:dns://8.8.8.8…
[02:33:57.436][127.0.0.1:64745] << CloseConnection({'connection': Server({'id': '…599044', 'address': ('8.8.8.8', 53), 'peername': ('8.8.8.8', 53), 'sockname': ('192.168.0.17', 64746), 'state': <ConnectionState.OPEN: 3>, 'transport_protocol': 'udp', 'timestamp_start': 1717196637.0141444})})
[02:33:57.436][127.0.0.1:64745] << CloseConnection({'connection': Server({'id': '…599044', 'address': ('8.8.8.8', 53), 'peername': ('8.8.8.8', 53), 'sockname': ('192.168.0.17', 64746), 'state': <ConnectionState.OPEN: 3>, 'transport_protocol': 'udp', 'timestamp_start': 1717196637.0141444}…
[02:33:57.437][127.0.0.1:64751] >> ConnectionClosed(connection=Client({'id': '…82296b', 'address': None, 'peername': ('127.0.0.1', 64751), 'sockname': ('0.0.0.0', 53), 'transport_protocol': 'udp', 'timestamp_start': 1717196637.3855908, 'proxy_mode': ProxyMode.parse('reverse:dns://8.8.8.8@53')}))
[02:33:57.437][127.0.0.1:64751] >> ConnectionClosed(connection=Client({'id': '…82296b', 'address': None, 'peername': ('127.0.0.1', 64751), 'sockname': ('0.0.0.0', 53), 'transport_protocol': 'udp', 'timestamp_start': 1717196637.3855908, 'proxy_mode': ProxyMode.parse('reverse:dns://8.8.8.8…
[02:33:57.437][127.0.0.1:64751] << CloseConnection({'connection': Server({'id': '…ef76b5', 'address': ('8.8.8.8', 53), 'peername': ('8.8.8.8', 53), 'sockname': ('192.168.0.17', 64752), 'state': <ConnectionState.OPEN: 3>, 'transport_protocol': 'udp', 'timestamp_start': 1717196637.3989928})})
[02:33:57.438][127.0.0.1:64751] << CloseConnection({'connection': Server({'id': '…ef76b5', 'address': ('8.8.8.8', 53), 'peername': ('8.8.8.8', 53), 'sockname': ('192.168.0.17', 64752), 'state': <ConnectionState.OPEN: 3>, 'transport_protocol': 'udp', 'timestamp_start': 1717196637.3989928}…
[02:33:57.438][127.0.0.1:64747] client disconnect
[02:33:57.439][127.0.0.1:64749] client disconnect
[02:33:57.439][127.0.0.1:64743] >> ConnectionClosed(connection=Server({'id': '…5eb969', 'address': ('8.8.8.8', 53), 'peername': ('8.8.8.8', 53), 'sockname': ('192.168.0.17', 64744), 'transport_protocol': 'udp', 'timestamp_start': 1717196636.908739}))
[02:33:57.440][127.0.0.1:64743] >> ConnectionClosed(connection=Server({'id': '…5eb969', 'address': ('8.8.8.8', 53), 'peername': ('8.8.8.8', 53), 'sockname': ('192.168.0.17', 64744), 'transport_protocol': 'udp', 'timestamp_start': 1717196636.908739}))
[02:33:57.443][127.0.0.1:64743] server disconnect 8.8.8.8:53
[02:33:57.443] UDP client task shutting down.
[02:33:57.444][127.0.0.1:64745] >> ConnectionClosed(connection=Server({'id': '…599044', 'address': ('8.8.8.8', 53), 'peername': ('8.8.8.8', 53), 'sockname': ('192.168.0.17', 64746), 'transport_protocol': 'udp', 'timestamp_start': 1717196637.0141444}))
[02:33:57.445][127.0.0.1:64745] >> ConnectionClosed(connection=Server({'id': '…599044', 'address': ('8.8.8.8', 53), 'peername': ('8.8.8.8', 53), 'sockname': ('192.168.0.17', 64746), 'transport_protocol': 'udp', 'timestamp_start': 1717196637.0141444}))
[02:33:57.445][127.0.0.1:64745] server disconnect 8.8.8.8:53
[02:33:57.446] UDP client task shutting down.
[02:33:57.446][127.0.0.1:64751] >> ConnectionClosed(connection=Server({'id': '…ef76b5', 'address': ('8.8.8.8', 53), 'peername': ('8.8.8.8', 53), 'sockname': ('192.168.0.17', 64752), 'transport_protocol': 'udp', 'timestamp_start': 1717196637.3989928}))
[02:33:57.447][127.0.0.1:64751] >> ConnectionClosed(connection=Server({'id': '…ef76b5', 'address': ('8.8.8.8', 53), 'peername': ('8.8.8.8', 53), 'sockname': ('192.168.0.17', 64752), 'transport_protocol': 'udp', 'timestamp_start': 1717196637.3989928}))
[02:33:57.447][127.0.0.1:64751] server disconnect 8.8.8.8:53
[02:33:57.447] UDP client task shutting down.
[02:33:57.448][127.0.0.1:64743] client disconnect
[02:33:57.448][127.0.0.1:64745] client disconnect
[02:33:57.449][127.0.0.1:64751] client disconnect
```
|
closed
|
2024-05-31T23:16:59Z
|
2024-09-30T21:38:33Z
|
https://github.com/mitmproxy/mitmproxy/issues/6886
|
[
"kind/bug",
"help wanted",
"area/rust"
] |
lostact
| 3
|
TracecatHQ/tracecat
|
fastapi
| 418
|
Empty action not picked up by validation (on commit)
|
**Describe the bug**
As described in title, results in unhelpful 500 internal server error in frontend, but in the API service logs you see the validation errors:
<img width="816" alt="Screenshot 2024-10-09 at 7 18 55 PM" src="https://github.com/user-attachments/assets/36290784-5a62-47a4-a37c-b0cd57e09b03">
**To Reproduce**
1. Create new workflow
2. Add reshape action
3. Press commit (do not save action)
4. Run workflow via manual trigger
**Expected behavior**
Error that action is empty on commit
**Screenshots**
<img width="1721" alt="Screenshot 2024-10-09 at 7 03 20 PM" src="https://github.com/user-attachments/assets/1056b841-f445-4a17-911d-6823ea191a49">
<img width="1303" alt="Screenshot 2024-10-09 at 7 03 10 PM" src="https://github.com/user-attachments/assets/761d416f-4d7e-467a-b4d9-d0a30b204be7">
|
closed
|
2024-10-10T02:19:13Z
|
2024-10-23T16:53:12Z
|
https://github.com/TracecatHQ/tracecat/issues/418
|
[
"bug",
"engine"
] |
topher-lo
| 0
|
sigmavirus24/github3.py
|
rest-api
| 258
|
User Keys Are Now Immutable
|
See: https://github.com/octokit/octokit.rb/pull/491
|
closed
|
2014-06-20T20:58:19Z
|
2014-12-03T04:42:03Z
|
https://github.com/sigmavirus24/github3.py/issues/258
|
[] |
sigmavirus24
| 0
|
CorentinJ/Real-Time-Voice-Cloning
|
python
| 1,106
|
Generated output audio is only 1 sec.
|
Unfortunately, I could not find a solution in closed issues, so I hope for help. The final audio is only 1 second. What could cause this to happen?
|
open
|
2022-09-04T13:49:03Z
|
2022-09-04T13:49:03Z
|
https://github.com/CorentinJ/Real-Time-Voice-Cloning/issues/1106
|
[] |
netrunner-exe
| 0
|
modAL-python/modAL
|
scikit-learn
| 62
|
Committee class does not update to new labels correctly.
|
Hi,
I encountered an exception in the `Committee` class when trying to capture the score after teaching the committee with unseen data containing new classes.
The problem seem to be that the `teach` function does the following:
1. Adds the new data to the training set X and y
2. Updates the known classes based on the classes known to the estimators given by the learners
3. Refits the estimators (hence updating the known classes to include the new classes)
Step 2 should not depend on the estimators or happen after Step 3.
I am working on a PR to fix this.
|
closed
|
2019-11-22T13:13:28Z
|
2019-11-26T08:17:17Z
|
https://github.com/modAL-python/modAL/issues/62
|
[] |
philipjhj
| 2
|
MaartenGr/BERTopic
|
nlp
| 1,447
|
Python 3.10 supported
|
Hi,
I'm wondering when will python 3.10 be supported?
Best,
|
open
|
2023-08-01T13:30:31Z
|
2023-08-01T17:23:46Z
|
https://github.com/MaartenGr/BERTopic/issues/1447
|
[] |
FlorentinLavaud
| 1
|
PaddlePaddle/PaddleHub
|
nlp
| 1,413
|
TypeError: the 'package' argument is required to perform a relative import for '.module'
|
欢迎您反馈PaddleHub使用问题,非常感谢您对PaddleHub的贡献!
在留下您的问题时,辛苦您同步提供如下信息:
- 版本、环境信息
1)PaddleHub和PaddlePaddle版本:PaddlePaddle版本为1.7.2,Paddlehub版本为1.6.2
2)系统环境:Windows,python3.7
- 复现信息:做cv模型的迁移学习
-代码如下: import paddlehub as hub
test_dir = './database/img.jpg'
model = hub.Module(directory='finetuned_model_to_module/')
prediction = model.predict(test_dir)
print(prediction)
-报错信息如下:Traceback (most recent call last):
File "E:/Data/AppData/firefox/demo2.py", line 4, in <module>
model = hub.Module(directory='finetuned_model_to_module/',use_gpu=False)
File "E:\miniconda3\lib\site-packages\paddlehub\module\module.py", line 104, in __new__
module = cls.init_with_directory(directory=directory, **kwargs)
File "E:\miniconda3\lib\site-packages\paddlehub\module\module.py", line 193, in init_with_directory
_module = importlib.import_module("{}.module".format(basename))
File "E:\miniconda3\lib\importlib\__init__.py", line 122, in import_module
raise TypeError(msg.format(name))
TypeError: the 'package' argument is required to perform a relative import for '.module'
文件夹下包含
1. finetuned_model_to_module
-module.py
-__init__.py
-model_finetune
2. demo2.py
|
closed
|
2021-05-14T09:03:20Z
|
2022-01-19T02:44:35Z
|
https://github.com/PaddlePaddle/PaddleHub/issues/1413
|
[] |
poopit
| 2
|
AUTOMATIC1111/stable-diffusion-webui
|
pytorch
| 15,578
|
[Feature Request]: Add a scheduler pattern to the filename save options now that it has been removed from the sampler in 1.9.0
|
### Is there an existing issue for this?
- [X] I have searched the existing issues and checked the recent builds/commits
### What would your feature do ?
In previous versions of webui, using the [sampler] pattern in the "Images filename pattern" setting would include the scheduler in the output text. Since 1.9.0 and the separation of the scheduler from the sample in the UI, this no longer works.
For example, previously [sampler] would add "DPM++ SDE Karras" into your image filename. Now it will just add "DPM++ SDE".
The [custom filename wiki page](https://github.com/AUTOMATIC1111/stable-diffusion-webui/wiki/Custom-Images-Filename-Name-and-Subdirectory#patterns) does not list any new option to include the scheduler in the filename either.
### Proposed workflow
1. Add an appropriate pattern option e.g. [scheduler] to the "Images filename pattern" setting that will add the appropriate text to the filename.
2. Update the wiki documentation if the feature is added.
### Additional information
_No response_
|
closed
|
2024-04-20T12:56:28Z
|
2024-04-21T04:11:11Z
|
https://github.com/AUTOMATIC1111/stable-diffusion-webui/issues/15578
|
[
"enhancement"
] |
roamingfrog
| 1
|
tfranzel/drf-spectacular
|
rest-api
| 650
|
Feature Request: Support for custom versioning classes
|
I have an application that uses AcceptHeaderVersioning in production but it has been migrated from no versioning at all.
In order to not break existing clients, we have the following `REST_FRAMEWORK` settings:
~~~python
'DEFAULT_VERSION': '1',
'ALLOWED_VERSIONS': ('1', '2'),
~~~
Some endpoints are available in version 1 and 2 and some are only available in version 1 (or no version given) and if the client always sends version 2, it will receive version 1 for endpoints that only support version 1. (And yes, since you'll be asking, the plan is to migrate those version 1 views directly to version 3 in case we want to change them... So each client expects "their" api version.)
I've tried to migrate that application from drf-yasg to drf-spectacular, since we already use JSONSchema for some views so support for openapi 3 would be a definite win for us. (Our JSON Schemata are even displayed in the swagger ui with drf-spectacular, but at the end of the page, and not at all in redoc... I think some other issue points that out as well.)
Unfortunately setting `DEFAULT_VERSION` is not compatible with the fix for #637, as the OP has pointed out.
Since giving an Accept Header is pretty inconvient in the browser (in redoc as well as the browsable DRF API), we have a custom versioning class that accepts `QueryParameterVersioning` as well as `AcceptHeaderVersioning` although the latter is required in production. This just works in `drf_yasg` (although we had problems customising the documentation for other parts of the api which may be easier in drf-spectacular).
If I disable the custom class and just use `AcceptHeaderVersioning`, I can generate the correct schema for each version on the command line and in the schema view, if I monkey-patch `modify_media_types_for_versioning` to change the media type for the default version as well, however it's not clear to me how to switch versions in swagger or redoc. (But maybe I simply overlooked something.)
Also probably being able to override a workflow method on a subclass would be a cleaner approach than having to monkey patch a function in `plumbing.py`.
So what are your thoughts on this?
|
closed
|
2022-02-08T16:09:00Z
|
2022-02-16T16:03:35Z
|
https://github.com/tfranzel/drf-spectacular/issues/650
|
[] |
TauPan
| 10
|
coqui-ai/TTS
|
python
| 2,699
|
Error
|
### Describe the bug
**hey**
> while installing the TTS, the error encountered is : ERROR: Ignored the following versions that require a different python version: 0.0.10.2 Requires-Python >=3.6.0, <3.9; 0.0.10.3 Requires-Python >=3.6.0, <3.9; 0.0.11 Requires-Python >=3.6.0, <3.9; 0.0.12 Requires-Python >=3.6.0, <3.9; 0.0.13.1 Requires-Python >=3.6.0, <3.9; 0.0.13.2 Requires-Python >=3.6.0, <3.9; 0.0.14.1 Requires-Python >=3.6.0, <3.9; 0.0.15 Requires-Python >=3.6.0, <3.9; 0.0.15.1 Requires-Python >=3.6.0, <3.9; 0.0.9 Requires-Python >=3.6.0, <3.9; 0.0.9.1 Requires-Python >=3.6.0, <3.9; 0.0.9.2 Requires-Python >=3.6.0, <3.9; 0.0.9a10 Requires-Python >=3.6.0, <3.9; 0.0.9a9 Requires-Python >=3.6.0, <3.9; 0.1.0 Requires-Python >=3.6.0, <3.10; 0.1.1 Requires-Python >=3.6.0, <3.10; 0.1.2 Requires-Python >=3.6.0, <3.10; 0.1.3 Requires-Python >=3.6.0, <3.10; 0.10.0 Requires-Python >=3.7.0, <3.11; 0.10.1 Requires-Python >=3.7.0, <3.11; 0.10.2 Requires-Python >=3.7.0, <3.11; 0.11.0 Requires-Python >=3.7.0, <3.11; 0.11.1 Requires-Python >=3.7.0, <3.11; 0.12.0 Requires-Python >=3.7.0, <3.11; 0.13.0 Requires-Python >=3.7.0, <3.11; 0.13.1 Requires-Python >=3.7.0, <3.11; 0.13.2 Requires-Python >=3.7.0, <3.11; 0.13.3 Requires-Python >=3.7.0, <3.11; 0.14.0 Requires-Python >=3.7.0, <3.11; 0.14.2 Requires-Python >=3.7.0, <3.11; 0.14.3 Requires-Python >=3.7.0, <3.11; 0.2.0 Requires-Python >=3.6.0, <3.10; 0.2.1 Requires-Python >=3.6.0, <3.10; 0.2.2 Requires-Python >=3.6.0, <3.10; 0.3.0 Requires-Python >=3.6.0, <3.10; 0.3.1 Requires-Python >=3.6.0, <3.10; 0.4.0 Requires-Python >=3.6.0, <3.10; 0.4.1 Requires-Python >=3.6.0, <3.10; 0.4.2 Requires-Python >=3.6.0, <3.10; 0.5.0 Requires-Python >=3.6.0, <3.10; 0.6.0 Requires-Python >=3.6.0, <3.10; 0.6.1 Requires-Python >=3.6.0, <3.10; 0.6.2 Requires-Python >=3.6.0, <3.10; 0.7.0 Requires-Python >=3.7.0, <3.11; 0.7.1 Requires-Python >=3.7.0, <3.11; 0.8.0 Requires-Python >=3.7.0, <3.11; 0.9.0 Requires-Python >=3.7.0, <3.11
> ERROR: Could not find a version that satisfies the requirement TTS (from versions: none)
> ERROR: No matching distribution found for TTS
>
### To Reproduce
pip install TTS
current python version : Python 3.11.1
### Expected behavior
it should get installed correctly
### Logs
_No response_
### Environment
```shell
.....
```
### Additional context
_No response_
|
closed
|
2023-06-22T10:23:14Z
|
2023-06-25T08:07:03Z
|
https://github.com/coqui-ai/TTS/issues/2699
|
[
"bug"
] |
junaidbashir11
| 1
|
BeanieODM/beanie
|
pydantic
| 971
|
[BUG] DeleteRules.DELETE_LINKS is not working?
|
**Describe the bug**
DeleteRules.DELETE_LINKS is not working. Delete a document success but all linked document didn't
**To Reproduce**
```python
import asyncio
from typing import Optional
from beanie import BackLink, Document, Link, init_beanie
from beanie.odm.utils.encoder import Encoder
from motor.motor_asyncio import AsyncIOMotorClient
from pydantic import Field
from beanie import DeleteRules, WriteRules
class Person(Document):
name: str
age: int
cars: list[Link["Car"]] = Field(default_factory=list)
class Car(Document):
manufacturer: str
price: float
owner: Optional[BackLink[Person]] = Field(default=None, original_field="cars")
async def init():
client = AsyncIOMotorClient(
"mongodb://localhost:27017",
)
await init_beanie(database=client.test_db, document_models=[Person, Car])
p1 = Person(
name="John",
age=25,
)
p1.cars = [
Car(manufacturer="Toyota", price=10000),
Car(manufacturer="BMW", price=20000)
]
await p1.save(link_rule=WriteRules.WRITE)
# The above code runs fine
person = await Person.find_one(Person.name == "John")
print(person)
# Only person document is deleted, cars didn't
await person.delete(link_rule=DeleteRules.DELETE_LINKS)
asyncio.run(init())
```
**Expected behavior**
- Document and all linked document will be deleted
**Additional context**
- Maybe I misunderstood how it works. Can you explain it?
|
closed
|
2024-07-13T15:20:57Z
|
2024-09-22T02:41:49Z
|
https://github.com/BeanieODM/beanie/issues/971
|
[
"Stale"
] |
dinhtuanan
| 4
|
dnouri/nolearn
|
scikit-learn
| 112
|
Different ways to install the packet
|
Hello, I tried to install the packet using pip as you suggest in the docs but I have some internet access problems from the command windows so I can't install it. I also tried in the old fashion, that means download the .zip , unzip and run $ python setup.py install but it doesn' work. Any other idea? Thank you
|
closed
|
2015-06-08T08:30:27Z
|
2015-06-15T12:27:55Z
|
https://github.com/dnouri/nolearn/issues/112
|
[] |
pattysan
| 3
|
open-mmlab/mmdetection
|
pytorch
| 11,832
|
多卡训练卡住,单卡GPU利用率为0,其余100%
|
如图:

自定义model后,训练总是出现卡住的情况。表现为时而单卡GPU利用率为0,其余皆为100%,训练速度大幅降低
而训练config中配置好的模型就不会出现这个问题
已经尝试export NCCL_P2P_DISABLE=1, 调整num_workers的方案,都没有用
有没有知道该如何解决
|
open
|
2024-07-05T05:22:10Z
|
2025-03-07T06:05:55Z
|
https://github.com/open-mmlab/mmdetection/issues/11832
|
[] |
WangJian981002
| 3
|
polyaxon/traceml
|
data-visualization
| 14
|
Patch level went down?
|
Hi, I am trying to get `pandas-summary` working, and had version 0.0.41 installed. I ran into the issue fixed by #11/#12, so I tried to upgrade. It seems like the version was changed to 0.0.5 with https://github.com/mouradmourafiq/pandas-summary/commit/42227d51d8d458ebce7090db971259565fb6ccdf
When I try to upgrade to 0.0.5, pip picks up the 0.0.41 version since its patch level is considered greater than 0.0.5. I am somewhat new to the Python world, so I could be missing an easy way to do this, but wondering if the new version should be updated to 0.0.42 or 0.1.0 to better comply with [semver](https://semver.org/).
```
pip install pandas-summary==
Collecting pandas-summary==
Could not find a version that satisfies the requirement pandas-summary== (from versions: 0.0.3, 0.0.4, 0.0.5, 0.0.41)
No matching distribution found for pandas-summary==
```
For now my workaround is to use `pip install pandas-summary==0.0.5` to install the exact version that I want to use.
|
closed
|
2018-06-21T20:42:06Z
|
2021-11-21T18:17:09Z
|
https://github.com/polyaxon/traceml/issues/14
|
[] |
panozzaj
| 0
|
sherlock-project/sherlock
|
python
| 2,122
|
Do we really need to package Sherlock for various platforms?
|
With the recent efforts to package Sherlock, this question came to mind:
Do we really need to package Sherlock for various platforms?
I personally don't think it's necessary for a few reasons:
- It would add more complexity to the entire workflow.
- It would be necessary to maintain packaging on multiple different platforms.
- The conventional way of packaging Python projects probably already works well for most; More below.
I believe that any platform that has some way to install Python along with Pip can theoretically install the Sherlock project without any problems. Publishing it only on PyPI seems to be the best path.
This would also be easier to maintain, due to having a single point of origin.
Sherlock doesn't seem like the type of project that needs specific packaging for each platform at the moment. With the exception of installation on Termux environments, which apparently has some issues installing pandas; I talk more about this here: https://github.com/sherlock-project/sherlock/issues/1945#issuecomment-1850757406
@sdushantha @ppfeister What do you think of these thoughts?
|
closed
|
2024-05-13T19:18:43Z
|
2024-11-05T01:28:08Z
|
https://github.com/sherlock-project/sherlock/issues/2122
|
[
"question"
] |
matheusfelipeog
| 17
|
graphdeco-inria/gaussian-splatting
|
computer-vision
| 1,019
|
Failed to build diff_gaussian_rasterization simple_knn
|
Hi there. Following [this video's](https://www.youtube.com/watch?v=UXtuigy_wYc) tutorial for Gaussian splatting and I get stuck in the `conda env create --file environment.yml` part. Throwing an error which is due to simple-knn
Error:
```
(base) C:\Users\rohil\gaussian-splatting>SET DISTUTILS_USE_SDK=1
(base) C:\Users\rohil\gaussian-splatting>conda env create --file environment.yml
Could not load conda plugin `anaconda-cloud-auth`:
cannot import name 'ChannelAuthBase' from 'conda.plugins.types' (C:\Users\rohil\anaconda3\lib\site-packages\conda\plugins\types.py)
WARNING conda.plugins.manager:load_entrypoints(84): Could not load conda plugin `anaconda-cloud-auth`:
cannot import name 'ChannelAuthBase' from 'conda.plugins.types' (C:\Users\rohil\anaconda3\lib\site-packages\conda\plugins\types.py)
Collecting package metadata (repodata.json): done
Solving environment: done
==> WARNING: A newer version of conda exists. <==
current version: 23.3.1
latest version: 24.9.2
Please update conda by running
$ conda update -n base -c defaults conda
Or to minimize the number of packages updated during conda update use
conda install conda=24.9.2
Downloading and Extracting Packages
Preparing transaction: done
Verifying transaction: done
Executing transaction: - "By downloading and using the CUDA Toolkit conda packages, you accept the terms and conditions of the CUDA End User License Agreement (EULA): https://docs.nvidia.com/cuda/eula/index.html"
done
Installing pip dependencies: / Ran pip subprocess with arguments:
['C:\\Users\\rohil\\anaconda3\\envs\\gaussian_splatting\\python.exe', '-m', 'pip', 'install', '-U', '-r', 'C:\\Users\\rohil\\gaussian-splatting\\condaenv.pry8d3d7.requirements.txt', '--exists-action=b']
Pip subprocess output:
Processing c:\users\rohil\gaussian-splatting\submodules\diff-gaussian-rasterization
Preparing metadata (setup.py): started
Preparing metadata (setup.py): finished with status 'done'
Processing c:\users\rohil\gaussian-splatting\submodules\simple-knn
Preparing metadata (setup.py): started
Preparing metadata (setup.py): finished with status 'done'
Building wheels for collected packages: diff_gaussian_rasterization, simple_knn
Building wheel for diff_gaussian_rasterization (setup.py): started
Building wheel for diff_gaussian_rasterization (setup.py): finished with status 'error'
Running setup.py clean for diff_gaussian_rasterization
Building wheel for simple_knn (setup.py): started
Building wheel for simple_knn (setup.py): finished with status 'error'
Running setup.py clean for simple_knn
Failed to build diff_gaussian_rasterization simple_knn
Installing collected packages: simple_knn, diff_gaussian_rasterization
Running setup.py install for simple_knn: started
Running setup.py install for simple_knn: finished with status 'error'
Pip subprocess error:
error: subprocess-exited-with-error
× python setup.py bdist_wheel did not run successfully.
│ exit code: 1
╰─> [89 lines of output]
running bdist_wheel
running build
running build_py
creating build
creating build\lib.win-amd64-cpython-37
creating build\lib.win-amd64-cpython-37\diff_gaussian_rasterization
copying diff_gaussian_rasterization\__init__.py -> build\lib.win-amd64-cpython-37\diff_gaussian_rasterization
running build_ext
C:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\torch\utils\cpp_extension.py:346: UserWarning: Error checking compiler version for cl: [WinError 2] The system cannot find the file specified
warnings.warn(f'Error checking compiler version for {compiler}: {error}')
C:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\torch\utils\cpp_extension.py:813: UserWarning: The detected CUDA version (11.8) has a minor version mismatch with the version that was used to compile PyTorch (11.6). Most likely this shouldn't be a problem.
warnings.warn(CUDA_MISMATCH_WARN.format(cuda_str_version, torch.version.cuda))
building 'diff_gaussian_rasterization._C' extension
creating C:\Users\rohil\gaussian-splatting\submodules\diff-gaussian-rasterization\build\temp.win-amd64-cpython-37
creating C:\Users\rohil\gaussian-splatting\submodules\diff-gaussian-rasterization\build\temp.win-amd64-cpython-37\Release
creating C:\Users\rohil\gaussian-splatting\submodules\diff-gaussian-rasterization\build\temp.win-amd64-cpython-37\Release\cuda_rasterizer
Emitting ninja build file C:\Users\rohil\gaussian-splatting\submodules\diff-gaussian-rasterization\build\temp.win-amd64-cpython-37\Release\build.ninja...
Compiling objects...
Allowing ninja to set a default number of workers... (overridable by setting the environment variable MAX_JOBS=N)
[1/5] cl /showIncludes /nologo /O2 /W3 /GL /DNDEBUG /MD /MD /wd4819 /wd4251 /wd4244 /wd4267 /wd4275 /wd4018 /wd4190 /EHsc -IC:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\torch\include -IC:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\torch\include\torch\csrc\api\include -IC:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\torch\include\TH -IC:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\torch\include\THC "-IC:\Program Files\NVIDIA GPU Computing Toolkit\CUDA\v11.8\include" -IC:\Users\rohil\anaconda3\envs\gaussian_splatting\include -IC:\Users\rohil\anaconda3\envs\gaussian_splatting\Include -c C:\Users\rohil\gaussian-splatting\submodules\diff-gaussian-rasterization\ext.cpp /FoC:\Users\rohil\gaussian-splatting\submodules\diff-gaussian-rasterization\build\temp.win-amd64-cpython-37\Release\ext.obj -DTORCH_API_INCLUDE_EXTENSION_H -DTORCH_EXTENSION_NAME=_C -D_GLIBCXX_USE_CXX11_ABI=0 /std:c++14
FAILED: C:/Users/rohil/gaussian-splatting/submodules/diff-gaussian-rasterization/build/temp.win-amd64-cpython-37/Release/ext.obj
cl /showIncludes /nologo /O2 /W3 /GL /DNDEBUG /MD /MD /wd4819 /wd4251 /wd4244 /wd4267 /wd4275 /wd4018 /wd4190 /EHsc -IC:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\torch\include -IC:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\torch\include\torch\csrc\api\include -IC:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\torch\include\TH -IC:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\torch\include\THC "-IC:\Program Files\NVIDIA GPU Computing Toolkit\CUDA\v11.8\include" -IC:\Users\rohil\anaconda3\envs\gaussian_splatting\include -IC:\Users\rohil\anaconda3\envs\gaussian_splatting\Include -c C:\Users\rohil\gaussian-splatting\submodules\diff-gaussian-rasterization\ext.cpp /FoC:\Users\rohil\gaussian-splatting\submodules\diff-gaussian-rasterization\build\temp.win-amd64-cpython-37\Release\ext.obj -DTORCH_API_INCLUDE_EXTENSION_H -DTORCH_EXTENSION_NAME=_C -D_GLIBCXX_Uninja: fatal: ReadFile: The handle is invalid.
SE_CXX11_ABI=0 /std:c++14
CreateProcess failed: The system cannot find the file specified.
Traceback (most recent call last):
File "C:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\torch\utils\cpp_extension.py", line 1814, in _run_ninja_build
env=env)
File "C:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\subprocess.py", line 512, in run
output=stdout, stderr=stderr)
subprocess.CalledProcessError: Command '['ninja', '-v']' returned non-zero exit status 1.
The above exception was the direct cause of the following exception:
Traceback (most recent call last):
File "<string>", line 36, in <module>
File "<pip-setuptools-caller>", line 34, in <module>
File "C:\Users\rohil\gaussian-splatting\submodules\diff-gaussian-rasterization\setup.py", line 32, in <module>
'build_ext': BuildExtension
File "C:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\setuptools\__init__.py", line 103, in setup
return distutils.core.setup(**attrs)
File "C:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\setuptools\_distutils\core.py", line 185, in setup
return run_commands(dist)
File "C:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\setuptools\_distutils\core.py", line 201, in run_commands
dist.run_commands()
File "C:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\setuptools\_distutils\dist.py", line 969, in run_commands
self.run_command(cmd)
File "C:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\setuptools\dist.py", line 963, in run_command
super().run_command(command)
File "C:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\setuptools\_distutils\dist.py", line 988, in run_command
cmd_obj.run()
File "C:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\wheel\bdist_wheel.py", line 368, in run
self.run_command("build")
File "C:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\setuptools\_distutils\cmd.py", line 318, in run_command
self.distribution.run_command(command)
File "C:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\setuptools\dist.py", line 963, in run_command
super().run_command(command)
File "C:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\setuptools\_distutils\dist.py", line 988, in run_command
cmd_obj.run()
File "C:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\setuptools\_distutils\command\build.py", line 131, in run
self.run_command(cmd_name)
File "C:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\setuptools\_distutils\cmd.py", line 318, in run_command
self.distribution.run_command(command)
File "C:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\setuptools\dist.py", line 963, in run_command
super().run_command(command)
File "C:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\setuptools\_distutils\dist.py", line 988, in run_command
cmd_obj.run()
File "C:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\setuptools\command\build_ext.py", line 88, in run
_build_ext.run(self)
File "C:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\setuptools\_distutils\command\build_ext.py", line 345, in run
self.build_extensions()
File "C:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\torch\utils\cpp_extension.py", line 765, in build_extensions
build_ext.build_extensions(self)
File "C:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\setuptools\_distutils\command\build_ext.py", line 467, in build_extensions
self._build_extensions_serial()
File "C:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\setuptools\_distutils\command\build_ext.py", line 493, in _build_extensions_serial
self.build_extension(ext)
File "C:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\setuptools\command\build_ext.py", line 249, in build_extension
_build_ext.build_extension(self, ext)
File "C:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\setuptools\_distutils\command\build_ext.py", line 555, in build_extension
depends=ext.depends,
File "C:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\torch\utils\cpp_extension.py", line 747, in win_wrap_ninja_compile
with_cuda=with_cuda)
File "C:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\torch\utils\cpp_extension.py", line 1492, in _write_ninja_file_and_compile_objects
error_prefix='Error compiling objects for extension')
File "C:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\torch\utils\cpp_extension.py", line 1824, in _run_ninja_build
raise RuntimeError(message) from e
RuntimeError: Error compiling objects for extension
[end of output]
note: This error originates from a subprocess, and is likely not a problem with pip.
ERROR: Failed building wheel for diff_gaussian_rasterization
error: subprocess-exited-with-error
× python setup.py bdist_wheel did not run successfully.
│ exit code: 1
╰─> [84 lines of output]
running bdist_wheel
running build
running build_ext
C:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\torch\utils\cpp_extension.py:346: UserWarning: Error checking compiler version for cl: [WinError 2] The system cannot find the file specified
warnings.warn(f'Error checking compiler version for {compiler}: {error}')
C:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\torch\utils\cpp_extension.py:813: UserWarning: The detected CUDA version (11.8) has a minor version mismatch with the version that was used to compile PyTorch (11.6). Most likely this shouldn't be a problem.
warnings.warn(CUDA_MISMATCH_WARN.format(cuda_str_version, torch.version.cuda))
building 'simple_knn._C' extension
creating C:\Users\rohil\gaussian-splatting\submodules\simple-knn\build
creating C:\Users\rohil\gaussian-splatting\submodules\simple-knn\build\temp.win-amd64-cpython-37
creating C:\Users\rohil\gaussian-splatting\submodules\simple-knn\build\temp.win-amd64-cpython-37\Release
Emitting ninja build file C:\Users\rohil\gaussian-splatting\submodules\simple-knn\build\temp.win-amd64-cpython-37\Release\build.ninja...
Compiling objects...
Allowing ninja to set a default number of workers... (overridable by setting the environment variable MAX_JOBS=N)
[1/3] cl /showIncludes /nologo /O2 /W3 /GL /DNDEBUG /MD /MD /wd4819 /wd4251 /wd4244 /wd4267 /wd4275 /wd4018 /wd4190 /EHsc -IC:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\torch\include -IC:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\torch\include\torch\csrc\api\include -IC:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\torch\include\TH -IC:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\torch\include\THC "-IC:\Program Files\NVIDIA GPU Computing Toolkit\CUDA\v11.8\include" -IC:\Users\rohil\anaconda3\envs\gaussian_splatting\include -IC:\Users\rohil\anaconda3\envs\gaussian_splatting\Include -c C:\Users\rohil\gaussian-splatting\submodules\simple-knn\ext.cpp /FoC:\Users\rohil\gaussian-splatting\submodules\simple-knn\build\temp.win-amd64-cpython-37\Release\ext.obj /wd4624 -DTORCH_API_INCLUDE_EXTENSION_H -DTORCH_EXTENSION_NAME=_C -D_GLIBCXX_USE_CXX11_ABI=0 /std:c++14
FAILED: C:/Users/rohil/gaussian-splatting/submodules/simple-knn/build/temp.win-amd64-cpython-37/Release/ext.obj
cl /showIncludes /nologo /O2 /W3 /GL /DNDEBUG /MD /MD /wd4819 /wd4251 /wd4244 /wd4267 /wd4275 /wd4018 /wd4190 /EHsc -IC:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\torch\include -IC:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\torch\include\torch\csrc\api\include -IC:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\torch\include\TH -IC:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\torch\include\THC "-IC:\Program Files\NVIDIA GPU Computing Toolkit\CUDA\v11.8\include" -IC:\Users\rohil\anaconda3\envs\gaussian_splatting\include -IC:\Users\rohil\anaconda3\envs\gaussian_splatting\Include -c C:\Users\rohil\gaussian-splatting\submodules\simple-knn\ext.cpp /FoC:\Users\rohil\gaussian-splatting\submodules\simple-knn\build\temp.win-amd64-cpython-37\Release\ext.obj /wd4624 -DTORCH_API_INCLUDE_EXTENSION_H -DTORCH_EXTENSION_NAME=_C -D_GLIBCXX_USE_CXX11_ABI=0 /std:c++14
CreateProcess failed: The system cannot finninja: fatal: ReadFile: The handle is invalid.
d the file specified.
Traceback (most recent call last):
File "C:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\torch\utils\cpp_extension.py", line 1814, in _run_ninja_build
env=env)
File "C:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\subprocess.py", line 512, in run
output=stdout, stderr=stderr)
subprocess.CalledProcessError: Command '['ninja', '-v']' returned non-zero exit status 1.
The above exception was the direct cause of the following exception:
Traceback (most recent call last):
File "<string>", line 36, in <module>
File "<pip-setuptools-caller>", line 34, in <module>
File "C:\Users\rohil\gaussian-splatting\submodules\simple-knn\setup.py", line 33, in <module>
'build_ext': BuildExtension
File "C:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\setuptools\__init__.py", line 103, in setup
return distutils.core.setup(**attrs)
File "C:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\setuptools\_distutils\core.py", line 185, in setup
return run_commands(dist)
File "C:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\setuptools\_distutils\core.py", line 201, in run_commands
dist.run_commands()
File "C:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\setuptools\_distutils\dist.py", line 969, in run_commands
self.run_command(cmd)
File "C:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\setuptools\dist.py", line 963, in run_command
super().run_command(command)
File "C:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\setuptools\_distutils\dist.py", line 988, in run_command
cmd_obj.run()
File "C:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\wheel\bdist_wheel.py", line 368, in run
self.run_command("build")
File "C:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\setuptools\_distutils\cmd.py", line 318, in run_command
self.distribution.run_command(command)
File "C:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\setuptools\dist.py", line 963, in run_command
super().run_command(command)
File "C:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\setuptools\_distutils\dist.py", line 988, in run_command
cmd_obj.run()
File "C:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\setuptools\_distutils\command\build.py", line 131, in run
self.run_command(cmd_name)
File "C:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\setuptools\_distutils\cmd.py", line 318, in run_command
self.distribution.run_command(command)
File "C:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\setuptools\dist.py", line 963, in run_command
super().run_command(command)
File "C:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\setuptools\_distutils\dist.py", line 988, in run_command
cmd_obj.run()
File "C:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\setuptools\command\build_ext.py", line 88, in run
_build_ext.run(self)
File "C:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\setuptools\_distutils\command\build_ext.py", line 345, in run
self.build_extensions()
File "C:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\torch\utils\cpp_extension.py", line 765, in build_extensions
build_ext.build_extensions(self)
File "C:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\setuptools\_distutils\command\build_ext.py", line 467, in build_extensions
self._build_extensions_serial()
File "C:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\setuptools\_distutils\command\build_ext.py", line 493, in _build_extensions_serial
self.build_extension(ext)
File "C:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\setuptools\command\build_ext.py", line 249, in build_extension
_build_ext.build_extension(self, ext)
File "C:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\setuptools\_distutils\command\build_ext.py", line 555, in build_extension
depends=ext.depends,
File "C:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\torch\utils\cpp_extension.py", line 747, in win_wrap_ninja_compile
with_cuda=with_cuda)
File "C:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\torch\utils\cpp_extension.py", line 1492, in _write_ninja_file_and_compile_objects
error_prefix='Error compiling objects for extension')
File "C:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\torch\utils\cpp_extension.py", line 1824, in _run_ninja_build
raise RuntimeError(message) from e
RuntimeError: Error compiling objects for extension
[end of output]
note: This error originates from a subprocess, and is likely not a problem with pip.
ERROR: Failed building wheel for simple_knn
error: subprocess-exited-with-error
× Running setup.py install for simple_knn did not run successfully.
│ exit code: 1
╰─> [99 lines of output]
running install
C:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\setuptools\_distutils\cmd.py:66: SetuptoolsDeprecationWarning: setup.py install is deprecated.
!!
********************************************************************************
Please avoid running ``setup.py`` directly.
Instead, use pypa/build, pypa/installer or other
standards-based tools.
See https://blog.ganssle.io/articles/2021/10/setup-py-deprecated.html for details.
********************************************************************************
!!
self.initialize_options()
running build
running build_ext
C:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\torch\utils\cpp_extension.py:346: UserWarning: Error checking compiler version for cl: [WinError 2] The system cannot find the file specified
warnings.warn(f'Error checking compiler version for {compiler}: {error}')
C:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\torch\utils\cpp_extension.py:813: UserWarning: The detected CUDA version (11.8) has a minor version mismatch with the version that was used to compile PyTorch (11.6). Most likely this shouldn't be a problem.
warnings.warn(CUDA_MISMATCH_WARN.format(cuda_str_version, torch.version.cuda))
building 'simple_knn._C' extension
creating C:\Users\rohil\gaussian-splatting\submodules\simple-knn\build
creating C:\Users\rohil\gaussian-splatting\submodules\simple-knn\build\temp.win-amd64-cpython-37
creating C:\Users\rohil\gaussian-splatting\submodules\simple-knn\build\temp.win-amd64-cpython-37\Release
Emitting ninja build file C:\Users\rohil\gaussian-splatting\submodules\simple-knn\build\temp.win-amd64-cpython-37\Release\build.ninja...
Compiling objects...
Allowing ninja to set a default number of workers... (overridable by setting the environment variable MAX_JOBS=N)
[1/3] cl /showIncludes /nologo /O2 /W3 /GL /DNDEBUG /MD /MD /wd4819 /wd4251 /wd4244 /wd4267 /wd4275 /wd4018 /wd4190 /EHsc -IC:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\torch\include -IC:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\torch\include\torch\csrc\api\include -IC:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\torch\include\TH -IC:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\torch\include\THC "-IC:\Program Files\NVIDIA GPU Computing Toolkit\CUDA\v11.8\include" -IC:\Users\rohil\anaconda3\envs\gaussian_splatting\include -IC:\Users\rohil\anaconda3\envs\gaussian_splatting\Include -c C:\Users\rohil\gaussian-splatting\submodules\simple-knn\ext.cpp /FoC:\Users\rohil\gaussian-splatting\submodules\simple-knn\build\temp.win-amd64-cpython-37\Release\ext.obj /wd4624 -DTORCH_API_INCLUDE_EXTENSION_H -DTORCH_EXTENSION_NAME=_C -D_GLIBCXX_USE_CXX11_ABI=0 /std:c++14
FAILED: C:/Users/rohil/gaussian-splatting/submodules/simple-knn/build/temp.win-amd64-cpython-37/Release/ext.obj
cl /showIncludes /nologo /O2 /W3 /GL /DNDEBUG /MD /MD /wd4819 /wd4251 /wd4244 /wd4267 /wd4275 /wd4018 /wd4190 /EHsc -IC:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\torch\include -IC:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\torch\include\torch\csrc\api\include -IC:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\torch\include\TH -IC:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\torch\include\THC "-IC:\Program Files\NVIDIA GPU Computing Toolkit\CUDA\v11.8\include" -IC:\Users\rohil\anaconda3\envs\gaussian_splatting\include -IC:\Users\rohil\anaconda3\envs\gaussian_splatting\Include -c C:\Users\rohil\gaussian-splatting\submodules\simple-knn\ext.cpp /FoC:\Users\rohil\gaussian-splatting\submodules\simple-knn\build\temp.win-amd64-cpython-37\Release\ext.obj /wd4624 -DTORCH_API_INCLUDE_EXTENSION_H -DTORCH_EXTENSION_NAME=_C -D_GLIBCXX_USE_CXX11_ABI=0 /std:c++14
CreateProcess failed: The system cannot finninja: fatal: ReadFile: The handle is invalid.
d the file specified.
Traceback (most recent call last):
File "C:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\torch\utils\cpp_extension.py", line 1814, in _run_ninja_build
env=env)
File "C:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\subprocess.py", line 512, in run
output=stdout, stderr=stderr)
subprocess.CalledProcessError: Command '['ninja', '-v']' returned non-zero exit status 1.
The above exception was the direct cause of the following exception:
Traceback (most recent call last):
File "<string>", line 36, in <module>
File "<pip-setuptools-caller>", line 34, in <module>
File "C:\Users\rohil\gaussian-splatting\submodules\simple-knn\setup.py", line 33, in <module>
'build_ext': BuildExtension
File "C:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\setuptools\__init__.py", line 103, in setup
return distutils.core.setup(**attrs)
File "C:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\setuptools\_distutils\core.py", line 185, in setup
return run_commands(dist)
File "C:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\setuptools\_distutils\core.py", line 201, in run_commands
dist.run_commands()
File "C:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\setuptools\_distutils\dist.py", line 969, in run_commands
self.run_command(cmd)
File "C:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\setuptools\dist.py", line 963, in run_command
super().run_command(command)
File "C:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\setuptools\_distutils\dist.py", line 988, in run_command
cmd_obj.run()
File "C:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\setuptools\command\install.py", line 78, in run
return orig.install.run(self)
File "C:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\setuptools\_distutils\command\install.py", line 697, in run
self.run_command('build')
File "C:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\setuptools\_distutils\cmd.py", line 318, in run_command
self.distribution.run_command(command)
File "C:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\setuptools\dist.py", line 963, in run_command
super().run_command(command)
File "C:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\setuptools\_distutils\dist.py", line 988, in run_command
cmd_obj.run()
File "C:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\setuptools\_distutils\command\build.py", line 131, in run
self.run_command(cmd_name)
File "C:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\setuptools\_distutils\cmd.py", line 318, in run_command
self.distribution.run_command(command)
File "C:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\setuptools\dist.py", line 963, in run_command
super().run_command(command)
File "C:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\setuptools\_distutils\dist.py", line 988, in run_command
cmd_obj.run()
File "C:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\setuptools\command\build_ext.py", line 88, in run
_build_ext.run(self)
File "C:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\setuptools\_distutils\command\build_ext.py", line 345, in run
self.build_extensions()
File "C:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\torch\utils\cpp_extension.py", line 765, in build_extensions
build_ext.build_extensions(self)
File "C:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\setuptools\_distutils\command\build_ext.py", line 467, in build_extensions
self._build_extensions_serial()
File "C:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\setuptools\_distutils\command\build_ext.py", line 493, in _build_extensions_serial
self.build_extension(ext)
File "C:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\setuptools\command\build_ext.py", line 249, in build_extension
_build_ext.build_extension(self, ext)
File "C:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\setuptools\_distutils\command\build_ext.py", line 555, in build_extension
depends=ext.depends,
File "C:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\torch\utils\cpp_extension.py", line 747, in win_wrap_ninja_compile
with_cuda=with_cuda)
File "C:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\torch\utils\cpp_extension.py", line 1492, in _write_ninja_file_and_compile_objects
error_prefix='Error compiling objects for extension')
File "C:\Users\rohil\anaconda3\envs\gaussian_splatting\lib\site-packages\torch\utils\cpp_extension.py", line 1824, in _run_ninja_build
raise RuntimeError(message) from e
RuntimeError: Error compiling objects for extension
[end of output]
note: This error originates from a subprocess, and is likely not a problem with pip.
error: legacy-install-failure
× Encountered error while trying to install package.
╰─> simple_knn
note: This is an issue with the package mentioned above, not pip.
hint: See above for output from the failure.
failed
CondaEnvException: Pip failed
```
I have tried some methods from the other issues like [this](https://github.com/graphdeco-inria/gaussian-splatting/issues/1014#issuecomment-2421791642) however to no avail.
Help is greatly appreciated.
Thanks!
|
closed
|
2024-10-20T15:01:08Z
|
2024-10-24T09:24:37Z
|
https://github.com/graphdeco-inria/gaussian-splatting/issues/1019
|
[] |
Ujjwal-Rohilla
| 1
|
BayesWitnesses/m2cgen
|
scikit-learn
| 287
|
NotImplementedError: Model 'numpy_ndarray' is not supported
|
while running `m2cgen --language java ModelOnline.joblib > model1.java` got error `NotImplementedError: Model 'numpy_ndarray' is not supported`. Are there plans to support it?
|
closed
|
2020-08-03T17:14:29Z
|
2020-08-04T15:56:30Z
|
https://github.com/BayesWitnesses/m2cgen/issues/287
|
[] |
yarix
| 5
|
unit8co/darts
|
data-science
| 2,743
|
[BUG] RIN and Likelihood models
|
**Describe the bug**
When using a Torch-based model with `use_reversible_instance_norm=True` and a `likelihood` specified (e.g., GaussianLikelihood), the model's predicted distribution parameters appear to be denormalized by the Reversible Instance Normalization (RINorm). This seems unintended, as RINorm should generally only affect the forecasted target values, not learned distribution parameters.
**To Reproduce**
```python
from darts.models import NLinearModel
from darts.utils.likelihood_models import GaussianLikelihood
# Constant TS creation
constant_two = np.ones(100) * 2
constant_two_series = TimeSeries.from_values(constant_two)
# Create normal model without likelihood and with reversible instance norm
model_normal = NLinearModel(20, 10, use_reversible_instance_norm=False)
model.fit(constant_two_series)
normal_pred_series = model.predict(20, series=constant_two_series)
# Create model with likelihood and with reversible instance norm
model_likelihood_with_revin = NLinearModel(20, 10, use_reversible_instance_norm=True, likelihood=GaussianLikelihood())
model_likelihood_with_revin.fit(constant_two_series)
likelihood_with_revin_pred_series = model_likelihood_with_revin.predict(20, series=constant_two_series)
# Create model with likelihood and with reversible instance norm
model_likelihood = NLinearModel(20, 10, use_reversible_instance_norm=False, likelihood=GaussianLikelihood())
model_likelihood.fit(constant_two_series)
likelihood_pred_series = model_likelihood.predict(20, series=constant_two_series)
# Create side-by-side plots
fig, axes = plt.subplots(1, 3, figsize=(14, 5))
# Left plot: Normal model
constant_two_series.plot(ax=axes[0], label="constant")
normal_pred_series.plot(ax=axes[0], label="normal")
axes[0].set_title("NLinear (no likelihood, with RevIN)")
axes[0].legend()
# Right plot: Likelihood model
constant_two_series.plot(ax=axes[1], label="constant")
likelihood_with_revin_pred_series.plot(ax=axes[1], label="likelihood")
axes[1].set_title("NLinear (with Gaussian Likelihood, with RevIN)")
axes[1].legend()
# Right plot: Likelihood model
constant_two_series.plot(ax=axes[2], label="constant")
likelihood_pred_series.plot(ax=axes[2], label="likelihood")
axes[2].set_title("NLinear (with Gaussian Likelihood, without RevIN)")
axes[2].legend()
plt.tight_layout()
plt.show()
```

**Expected behavior**
While denormalizing the predicted output values makes sense in standard forecasting scenarios, applying this
enormalization to distribution parameters (like the mean or variance from a likelihood model) is typically not appropriate.
These parameters should remain in the normalized space unless explicitly required otherwise.
**System (please complete the following information):**
- Python version: 3.11.11
- darts version: 0.34.0
**Additional context**
When inspecting the code responsible for applying RINorm—specifically in `darts.models.forecasting.pl_forecasting_module`, lines 48 to 67—it appears that all outputs of the model are denormalized.
[Link to Code](https://github.com/unit8co/darts/blob/42776790183bbe42411fc2dba3e1e8416f9263e8/darts/models/forecasting/pl_forecasting_module.py#L48C5-L67C27)
|
open
|
2025-03-24T08:13:58Z
|
2025-03-24T08:19:43Z
|
https://github.com/unit8co/darts/issues/2743
|
[
"bug",
"triage"
] |
PaulRabich
| 0
|
wagtail/wagtail
|
django
| 12,210
|
The Workflow task type filter doesn't work in the task chooser modal
|
To reproduce:
- add a custom workflow task
- create said task type
- edit a workflow to add the task and choose from existing
https://www.loom.com/share/c9db3cac043a47938dd862655e9764a9?sid=33905af7-34c2-4419-bfb1-1ab598a66c25
Wagtail 6.2+
Confirmed it works in Wagtail 6.1
|
closed
|
2024-08-06T14:10:47Z
|
2024-08-08T17:52:30Z
|
https://github.com/wagtail/wagtail/issues/12210
|
[
"type:Bug",
"component:Workflow"
] |
zerolab
| 1
|
aiortc/aiortc
|
asyncio
| 257
|
Videostream-cli webcam to another videostream-cli not working
|
Thank you for this amazing package.
I am trying to use this package for streaming on raspberry pi to Ubuntu machine receiving frames and handle it with opencv like in server example.
Tried those on raspberry pi:
1. Server example works fine
2. Webcam works fine
3. Videostream-cli example, streaming an mp4 video works fine and timing is correct
But running videostream-cli with webcam instead of video doesn't work, I receive first one or couple of frames correctly then the stream get stopped hang or receiving curropted frames.
* I had tried different type of cameras
* and test on raspberry pi 3 and 4
* tried different versions of ffmpeg even downloading source and compile it
Non of them was able to stream successfully.
Why is this happening since both sever and webcam both works fine, means PYAV and FFMPEG installed correctly.
This how I am running videostream-cli:
on raspberry pi:
`python cli.py offer`
and this how I modified the code to make offer stream from webcam:
```
# create media source
if args.play_from:
player = MediaPlayer(args.play_from)
else:
if args.role == "offer":
options = {"framerate": "30", "video_size": "640x480"}
player = MediaPlayer("/dev/video0", format="v4l2", options=options)
else:
player = None
````
on Ubuntu machine I run
`python cli.py answer --record-to video.mp4`
|
closed
|
2020-01-26T17:14:18Z
|
2021-03-07T14:53:25Z
|
https://github.com/aiortc/aiortc/issues/257
|
[
"question"
] |
AnasMK
| 8
|
jupyter/docker-stacks
|
jupyter
| 1,852
|
[BUG] - file permission problem on CentOS 7
|
### What docker image(s) are you using?
base-notebook
### OS system and architecture running docker image
CentOS 7.6.1810 / amd64
### What Docker command are you running?
'docker run -d --restart=always --name=jupyterlab -p 18888:8888 jupyter/base-notebook'
### How to Reproduce the problem?
1. Run the docker run command.
2. Open the jupyter url, it could be opened, but the file browser is blank. Error information is in the container logs.
3. Try to create a notebook, it will display: Error Unexpected error while saving file: Untitled1.ipynb [Errno 2] No such file or directory: '/home/jovyan/.~Untitled1.ipynb' -> '/home/jovyan/Untitled1.ipynb'
### Command output
```bash session
#before access the url:
$ docker logs jupyterlab
Entered start.sh with args: jupyter lab
WARNING: no write access to /home/jovyan. Try starting the container with group 'users' (100), e.g. using "--group-add=users".
Executing the command: jupyter lab
Fail to get yarn configuration. /opt/conda/bin/node[15]: ../../src/node_platform.cc:61:std::unique_ptr<long unsigned int> node::WorkerThreadsTaskRunner::DelayedTaskScheduler::Start(): Assertion `(0) == (uv_thread_create(t.get(), start_thread, this))' failed.
1: 0x7fb61bcec5e9 node::Abort() [/opt/conda/bin/../lib/libnode.so.108]
2: 0x7fb61bcec67b [/opt/conda/bin/../lib/libnode.so.108]
3: 0x7fb61bd6254d node::WorkerThreadsTaskRunner::WorkerThreadsTaskRunner(int) [/opt/conda/bin/../lib/libnode.so.108]
4: 0x7fb61bd62632 node::NodePlatform::NodePlatform(int, v8::TracingController*, v8::PageAllocator*) [/opt/conda/bin/../lib/libnode.so.108]
5: 0x7fb61bcae367 [/opt/conda/bin/../lib/libnode.so.108]
6: 0x7fb61bcaee67 node::InitializeOncePerProcess(std::vector<std::__cxx11::basic_string<char, std::char_traits<char>, std::allocator<char> >, std::allocator<std::__cxx11::basic_string<char, std::char_traits<char>, std::allocator<char> > > > const&, node::ProcessFlags::Flags) [/opt/conda/bin/../lib/libnode.so.108]
7: 0x7fb61bcaf223 node::Start(int, char**) [/opt/conda/bin/../lib/libnode.so.108]
8: 0x7fb61b289d90 [/lib/x86_64-linux-gnu/libc.so.6]
9: 0x7fb61b289e40 __libc_start_main [/lib/x86_64-linux-gnu/libc.so.6]
10: 0x55947ac6507f [/opt/conda/bin/node]
[I 2022-12-28 09:24:05.743 ServerApp] jupyter_server_terminals | extension was successfully linked.
[I 2022-12-28 09:24:05.749 ServerApp] jupyterlab | extension was successfully linked.
[W 2022-12-28 09:24:05.752 NotebookApp] 'ip' has moved from NotebookApp to ServerApp. This config will be passed to ServerApp. Be sure to update your config before our next release.
[W 2022-12-28 09:24:05.752 NotebookApp] 'port' has moved from NotebookApp to ServerApp. This config will be passed to ServerApp. Be sure to update your config before our next release.
[W 2022-12-28 09:24:05.752 NotebookApp] 'port' has moved from NotebookApp to ServerApp. This config will be passed to ServerApp. Be sure to update your config before our next release.
[I 2022-12-28 09:24:05.755 ServerApp] nbclassic | extension was successfully linked.
[I 2022-12-28 09:24:05.757 ServerApp] Writing Jupyter server cookie secret to /home/jovyan/.local/share/jupyter/runtime/jupyter_cookie_secret
[I 2022-12-28 09:24:06.005 ServerApp] notebook_shim | extension was successfully linked.
[I 2022-12-28 09:24:06.348 ServerApp] notebook_shim | extension was successfully loaded.
[I 2022-12-28 09:24:06.350 ServerApp] jupyter_server_terminals | extension was successfully loaded.
[I 2022-12-28 09:24:06.351 LabApp] JupyterLab extension loaded from /opt/conda/lib/python3.10/site-packages/jupyterlab
[I 2022-12-28 09:24:06.351 LabApp] JupyterLab application directory is /opt/conda/share/jupyter/lab
[I 2022-12-28 09:24:06.355 ServerApp] jupyterlab | extension was successfully loaded.
[I 2022-12-28 09:24:06.366 ServerApp] nbclassic | extension was successfully loaded.
[I 2022-12-28 09:24:06.366 ServerApp] Serving notebooks from local directory: /home/jovyan
[I 2022-12-28 09:24:06.366 ServerApp] Jupyter Server 2.0.5 is running at:
[I 2022-12-28 09:24:06.366 ServerApp] http://5fbe0adaadef:8888/lab?token=6ffb6ebf104eb5eae301400b3142aa1c0dbc065ae3387a9d
[I 2022-12-28 09:24:06.366 ServerApp] or http://127.0.0.1:8888/lab?token=6ffb6ebf104eb5eae301400b3142aa1c0dbc065ae3387a9d
[I 2022-12-28 09:24:06.366 ServerApp] Use Control-C to stop this server and shut down all kernels (twice to skip confirmation).
[C 2022-12-28 09:24:06.370 ServerApp]
To access the server, open this file in a browser:
file:///home/jovyan/.local/share/jupyter/runtime/jpserver-8-open.html
Or copy and paste one of these URLs:
http://5fbe0adaadef:8888/lab?token=6ffb6ebf104eb5eae301400b3142aa1c0dbc065ae3387a9d
or http://127.0.0.1:8888/lab?token=6ffb6ebf104eb5eae301400b3142aa1c0dbc065ae3387a9d
```
### Expected behavior
_No response_
### Actual behavior
It can not be used at all.
### Anything else?
The funny part is, when I buildt docker-stacks-foundation and base-notebook images myself, only changing the ROOT_CONTAINER from ubuntu:22.04 to ubuntu:focal in the Dockerfile, the problem disappeard. It seems that the Docker image ubuntu:22.04 has file permission problem with CentOS 7. I tried many other methods on adapting the file permissions, they just didn't work.
The log is here.
[jupyterlab.log](https://github.com/jupyter/docker-stacks/files/10313412/jupyterlab.log)
|
closed
|
2022-12-28T09:53:03Z
|
2022-12-28T20:17:22Z
|
https://github.com/jupyter/docker-stacks/issues/1852
|
[
"type:Bug"
] |
iskoldt-X
| 1
|
plotly/dash
|
plotly
| 3,138
|
Bump Flask and Werkzeug versions
|
We pin `flask` and `werkzeug` to older (3.0.6 / 3.0.3) versions. We should upgrade these.
We'll need to ensure compatibility across our ecosystem (OSS and Enterprise libraries). This is likely not something for 3.0 but worth considering.
|
open
|
2025-01-29T14:07:00Z
|
2025-03-11T14:17:10Z
|
https://github.com/plotly/dash/issues/3138
|
[
"infrastructure",
"P2"
] |
ndrezn
| 2
|
davidsandberg/facenet
|
computer-vision
| 760
|
how to determine the model has convergence when train the model?
|
Is it cost a lot of time to make the model convergence?
|
open
|
2018-05-25T07:15:23Z
|
2018-05-25T07:15:23Z
|
https://github.com/davidsandberg/facenet/issues/760
|
[] |
boyliwensheng
| 0
|
junyanz/pytorch-CycleGAN-and-pix2pix
|
pytorch
| 686
|
Any reason can cause color blocks?
|

when i use pix2pix train the model, it can generate the picture with color blocks, anyone can help me what reason may cause this problem, thanks.
|
closed
|
2019-06-28T03:57:52Z
|
2020-07-28T20:26:34Z
|
https://github.com/junyanz/pytorch-CycleGAN-and-pix2pix/issues/686
|
[] |
wadesunyang
| 1
|
keras-team/keras
|
tensorflow
| 20,063
|
Dropout in ConvLSTM Cell
|
Really quick question here.
In the ConvLSTMCell [here](https://github.com/keras-team/keras/blob/v3.3.3/keras/src/layers/rnn/conv_lstm.py#L236-L242), the dropout code has been commented out. `recurrent_dropout` also doesn't seem to do anything, except it is used in the `DropoutRNNCell` mixin.
Is this a bug or am I missing something?
|
closed
|
2024-07-30T00:44:05Z
|
2024-08-06T16:46:55Z
|
https://github.com/keras-team/keras/issues/20063
|
[
"type:Bug"
] |
dryglicki
| 5
|
Kanaries/pygwalker
|
pandas
| 24
|
Embedding directly in html
|
So I would love to use pygwalker in my project which is currently serving the data analytics through a simple flask server. Is there any easy way already available?
|
closed
|
2023-02-22T09:32:43Z
|
2023-02-24T08:34:09Z
|
https://github.com/Kanaries/pygwalker/issues/24
|
[] |
DeastinY
| 4
|
mwaskom/seaborn
|
matplotlib
| 3,701
|
Feature Request: Continuous axes heat map
|
Feature Request:
Continuous axes heat map.
This would function similarly to the existing heatmap feature but allow for continuous axes rather than purely categorical.
On the backend, it would behave more similarly to a 2d histplot, but instead of performing a count of data the function would accept an array_like containing values or perhaps keywords corresponding to aggregators (e.g. 'min', 'max', etc.). A special case would be 'count' which would behave like a regular histogram.
Many thanks for your excellent work maintaining an excellent library.
|
closed
|
2024-05-31T03:55:38Z
|
2025-01-26T15:39:56Z
|
https://github.com/mwaskom/seaborn/issues/3701
|
[] |
HThawley
| 1
|
kizniche/Mycodo
|
automation
| 1,091
|
Dashboard not displaying after 8.12.6 upgrade
|
### Describe the problem/bug
After upgrading to the latest version v8.12.6 the Dashboard is no longer dispalying correctly - just empty boxes. Issue on Chrome (desktop) and Safari (phone).
I've tried adding and editing widgets to no avail. Seems to be an issue with pulling the data?
### Versions:
- Mycodo Version: 8.12.6
- Raspberry Pi Version: 3B+
- Raspbian OS Version: Buster
### Screenshots
Thanks for the awesome work that you are doing with Mycodo. I've been using it for the past 5 years for growing mushrooms. I see you recently started a new project - really cool!
|
closed
|
2021-09-17T21:41:09Z
|
2021-09-18T06:43:55Z
|
https://github.com/kizniche/Mycodo/issues/1091
|
[] |
jacopienaar
| 9
|
ResidentMario/missingno
|
data-visualization
| 157
|
Value Error while using missingno bar
|
While working on a kaggle dataset for june challenge i came across this error.
ValueError: The number of FixedLocator locations (0), usually from a call to set_ticks, does not match the number of ticklabels (81).
I am using this code
import missingno as msno
plt.figure(figsize = (20,10))
msno.bar(data)
plt.show()
Please fix the issue.
|
closed
|
2022-06-10T14:00:07Z
|
2022-11-20T08:48:10Z
|
https://github.com/ResidentMario/missingno/issues/157
|
[] |
prashant2-4-4
| 1
|
huggingface/datasets
|
pandas
| 7,326
|
Remove upper bound for fsspec
|
### Describe the bug
As also raised by @cyyever in https://github.com/huggingface/datasets/pull/7296 and @NeilGirdhar in https://github.com/huggingface/datasets/commit/d5468836fe94e8be1ae093397dd43d4a2503b926#commitcomment-140952162 , `datasets` has a problematic version constraint on `fsspec`.
In our case this causes (unnecessary?) troubles due to a race condition bug in that version of the corresponding `gcsfs` plugin, that causes deadlocks: https://github.com/fsspec/gcsfs/pull/643
We just use a version override to ignore the constraint from `datasets`, but imho the version constraint could just be removed in the first place?
The last few PRs bumping the upper bound were basically uneventful:
* https://github.com/huggingface/datasets/pull/7219
* https://github.com/huggingface/datasets/pull/6921
* https://github.com/huggingface/datasets/pull/6747
### Steps to reproduce the bug
-
### Expected behavior
Installing `fsspec>=2024.10.0` along `datasets` should be possible without overwriting constraints.
### Environment info
All recent datasets versions
|
open
|
2024-12-13T11:35:12Z
|
2025-01-03T15:34:37Z
|
https://github.com/huggingface/datasets/issues/7326
|
[] |
fellhorn
| 1
|
airtai/faststream
|
asyncio
| 1,986
|
Bug: FastAPI raised exception "AnyDict not defined" while adding RabbitMessage as parameter to subscriber
|
**Describe the bug**
I am trying to amend my current FastAPI based app with subscribers for RabbitMQ.
Inside the subscriber function I need access to the full message or at least the correlation_id and the headers. However, I seem not to get acces to them from the Request parameter, thus, trying to add the `message: RabbitMessage`, but then it explodes.
On the FastApi Lifespan `yield` this following exception is raised, but *only* if the subscriber endpoint ask for RabbitMessage:
```
File "/.../.venv/lib/python3.12/site-packages/pydantic/type_adapter.py", line 270, in _init_core_attrs
self._core_schema = _getattr_no_parents(self._type, '__pydantic_core_schema__')
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/.../.venv/lib/python3.12/site-packages/pydantic/type_adapter.py", line 112, in _getattr_no_parents
raise AttributeError(attribute)
AttributeError: __pydantic_core_schema__
During handling of the above exception, another exception occurred:
Traceback (most recent call last):
File "/.../.venv/lib/python3.12/site-packages/pydantic/_internal/_generate_schema.py", line 864, in _resolve_forward_ref
obj = _typing_extra.eval_type_backport(obj, globalns=self._types_namespace)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/.../.venv/lib/python3.12/site-packages/pydantic/_internal/_typing_extra.py", line 279, in eval_type_backport
return _eval_type_backport(value, globalns, localns, type_params)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/.../.venv/lib/python3.12/site-packages/pydantic/_internal/_typing_extra.py", line 303, in _eval_type_backport
return _eval_type(value, globalns, localns, type_params)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/.../.venv/lib/python3.12/site-packages/pydantic/_internal/_typing_extra.py", line 332, in _eval_type
return typing._eval_type( # type: ignore
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/usr/lib/python3.12/typing.py", line 414, in _eval_type
return t._evaluate(globalns, localns, recursive_guard)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/usr/lib/python3.12/typing.py", line 924, in _evaluate
eval(self.__forward_code__, globalns, localns),
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "<string>", line 1, in <module>
NameError: name 'AnyDict' is not defined
The above exception was the direct cause of the following exception:
Traceback (most recent call last):
File "/.../.venv/lib/python3.12/site-packages/starlette/routing.py", line 693, in lifespan
async with self.lifespan_context(app) as maybe_state:
File "/usr/lib/python3.12/contextlib.py", line 210, in __aenter__
return await anext(self.gen)
^^^^^^^^^^^^^^^^^^^^^
File "/.../.venv/lib/python3.12/site-packages/fastapi/routing.py", line 134, in merged_lifespan
async with nested_context(app) as maybe_nested_state:
File "/usr/lib/python3.12/contextlib.py", line 210, in __aenter__
return await anext(self.gen)
^^^^^^^^^^^^^^^^^^^^^
File "/.../.venv/lib/python3.12/site-packages/faststream/broker/fastapi/router.py", line 324, in start_broker_lifespan
await self.broker.start()
File "/.../.venv/lib/python3.12/site-packages/faststream/rabbit/broker/broker.py", line 508, in start
await super().start()
File "/.../.venv/lib/python3.12/site-packages/faststream/broker/core/usecase.py", line 219, in start
await self.connect()
File "/.../.venv/lib/python3.12/site-packages/faststream/rabbit/broker/broker.py", line 426, in connect
connection = await super().connect(**kwargs)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/.../.venv/lib/python3.12/site-packages/faststream/broker/core/usecase.py", line 227, in connect
self.setup()
File "/.../.venv/lib/python3.12/site-packages/faststream/broker/core/usecase.py", line 238, in setup
self.setup_subscriber(h)
File "/.../.venv/lib/python3.12/site-packages/faststream/broker/core/usecase.py", line 251, in setup_subscriber
subscriber.setup(**data)
File "/.../.venv/lib/python3.12/site-packages/faststream/rabbit/subscriber/usecase.py", line 127, in setup
super().setup(
File "/.../.venv/lib/python3.12/site-packages/faststream/broker/subscriber/usecase.py", line 182, in setup
call.setup(
File "/.../.venv/lib/python3.12/site-packages/faststream/broker/subscriber/call_item.py", line 92, in setup
dependant = self.handler.set_wrapped(
^^^^^^^^^^^^^^^^^^^^^^^^^
File "/.../.venv/lib/python3.12/site-packages/faststream/broker/wrapper/call.py", line 159, in set_wrapped
call = decor(call)
^^^^^^^^^^^
File "/.../.venv/lib/python3.12/site-packages/faststream/broker/fastapi/router.py", line 231, in wrapper
return wrap_callable_to_fastapi_compatible(
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/.../.venv/lib/python3.12/site-packages/faststream/broker/fastapi/route.py", line 94, in wrap_callable_to_fastapi_compatible
dependent=get_fastapi_native_dependant(user_callable, list(dependencies)),
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/.../.venv/lib/python3.12/site-packages/faststream/broker/fastapi/get_dependant.py", line 32, in get_fastapi_native_dependant
dependent = get_dependant(
^^^^^^^^^^^^^^
File "/.../.venv/lib/python3.12/site-packages/fastapi/dependencies/utils.py", line 277, in get_dependant
param_details = analyze_param(
^^^^^^^^^^^^^^
File "/.../.venv/lib/python3.12/site-packages/fastapi/dependencies/utils.py", line 478, in analyze_param
field = create_model_field(
^^^^^^^^^^^^^^^^^^^
File "/.../.venv/lib/python3.12/site-packages/fastapi/utils.py", line 96, in create_model_field
return ModelField(**kwargs) # type: ignore[arg-type]
^^^^^^^^^^^^^^^^^^^^
File "<string>", line 6, in __init__
File "/.../.venv/lib/python3.12/site-packages/fastapi/_compat.py", line 110, in __post_init__
self._type_adapter: TypeAdapter[Any] = TypeAdapter(
^^^^^^^^^^^^
File "/.../.venv/lib/python3.12/site-packages/pydantic/type_adapter.py", line 257, in __init__
self._init_core_attrs(rebuild_mocks=False)
File "/.../.venv/lib/python3.12/site-packages/pydantic/type_adapter.py", line 135, in wrapped
return func(self, *args, **kwargs)
^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/.../.venv/lib/python3.12/site-packages/pydantic/type_adapter.py", line 277, in _init_core_attrs
self._core_schema = _get_schema(self._type, config_wrapper, parent_depth=self._parent_depth)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/.../.venv/lib/python3.12/site-packages/pydantic/type_adapter.py", line 95, in _get_schema
schema = gen.generate_schema(type_)
^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/.../.venv/lib/python3.12/site-packages/pydantic/_internal/_generate_schema.py", line 655, in generate_schema
schema = self._generate_schema_inner(obj)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/.../.venv/lib/python3.12/site-packages/pydantic/_internal/_generate_schema.py", line 908, in _generate_schema_inner
return self._annotated_schema(obj)
^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/.../.venv/lib/python3.12/site-packages/pydantic/_internal/_generate_schema.py", line 2028, in _annotated_schema
schema = self._apply_annotations(source_type, annotations)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/.../.venv/lib/python3.12/site-packages/pydantic/_internal/_generate_schema.py", line 2107, in _apply_annotations
schema = get_inner_schema(source_type)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/.../.venv/lib/python3.12/site-packages/pydantic/_internal/_schema_generation_shared.py", line 83, in __call__
schema = self._handler(source_type)
^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/.../.venv/lib/python3.12/site-packages/pydantic/_internal/_generate_schema.py", line 2189, in new_handler
schema = metadata_get_schema(source, get_inner_schema)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/.../.venv/lib/python3.12/site-packages/pydantic/_internal/_generate_schema.py", line 2185, in <lambda>
lambda source, handler: handler(source)
^^^^^^^^^^^^^^^
File "/.../.venv/lib/python3.12/site-packages/pydantic/_internal/_schema_generation_shared.py", line 83, in __call__
schema = self._handler(source_type)
^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/.../.venv/lib/python3.12/site-packages/pydantic/_internal/_generate_schema.py", line 2189, in new_handler
schema = metadata_get_schema(source, get_inner_schema)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/.../.venv/lib/python3.12/site-packages/pydantic/_internal/_generate_schema.py", line 2185, in <lambda>
lambda source, handler: handler(source)
^^^^^^^^^^^^^^^
File "/.../.venv/lib/python3.12/site-packages/pydantic/_internal/_schema_generation_shared.py", line 83, in __call__
schema = self._handler(source_type)
^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/.../.venv/lib/python3.12/site-packages/pydantic/_internal/_generate_schema.py", line 2088, in inner_handler
schema = self._generate_schema_inner(obj)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/.../.venv/lib/python3.12/site-packages/pydantic/_internal/_generate_schema.py", line 929, in _generate_schema_inner
return self.match_type(obj)
^^^^^^^^^^^^^^^^^^^^
File "/.../.venv/lib/python3.12/site-packages/pydantic/_internal/_generate_schema.py", line 1025, in match_type
return self._dataclass_schema(obj, None)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/.../.venv/lib/python3.12/site-packages/pydantic/_internal/_generate_schema.py", line 1822, in _dataclass_schema
args = sorted(
^^^^^^^
File "/.../.venv/lib/python3.12/site-packages/pydantic/_internal/_generate_schema.py", line 1823, in <genexpr>
(self._generate_dc_field_schema(k, v, decorators) for k, v in fields.items()),
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/.../.venv/lib/python3.12/site-packages/pydantic/_internal/_generate_schema.py", line 1132, in _generate_dc_field_schema
common_field = self._common_field_schema(name, field_info, decorators)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/.../.venv/lib/python3.12/site-packages/pydantic/_internal/_generate_schema.py", line 1308, in _common_field_schema
schema = self._apply_annotations(
^^^^^^^^^^^^^^^^^^^^^^^^
File "/.../.venv/lib/python3.12/site-packages/pydantic/_internal/_generate_schema.py", line 2107, in _apply_annotations
schema = get_inner_schema(source_type)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/.../.venv/lib/python3.12/site-packages/pydantic/_internal/_schema_generation_shared.py", line 83, in __call__
schema = self._handler(source_type)
^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/.../.venv/lib/python3.12/site-packages/pydantic/_internal/_generate_schema.py", line 2088, in inner_handler
schema = self._generate_schema_inner(obj)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/.../.venv/lib/python3.12/site-packages/pydantic/_internal/_generate_schema.py", line 918, in _generate_schema_inner
return self.generate_schema(self._resolve_forward_ref(obj))
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/.../.venv/lib/python3.12/site-packages/pydantic/_internal/_generate_schema.py", line 866, in _resolve_forward_ref
raise PydanticUndefinedAnnotation.from_name_error(e) from e
pydantic.errors.PydanticUndefinedAnnotation: name 'AnyDict' is not defined
For further information visit https://errors.pydantic.dev/2.9/u/undefined-annotation
```
**How to reproduce**
Include source code:
The faststream subscriber setup:
```python
from fastapi import FastAPI, Request
from faststream.rabbit.fastapi import RabbitRouter
from faststream.rabbit import RabbitBroker, RabbitQueue, RabbitExchange
from faststream.rabbit import RabbitMessage
from faststream.types import AnyDict
@rabbit_router.subscriber(queue=action_queue, no_ack=False, retry=3, include_in_schema=False)
async def action_subscriber(request: Request, body: str, message: RabbitMessage):
logger.info(f"Action subscriber received message: {body}")
await publish_message(f"Action subscriber received message: {body}", exchange="logs", sub_channel='info')
```
The App.py.
```python
from fastapi import FastAPI, status, Request
...
def create_app() -> FastAPI:
@asynccontextmanager
async def lifespan(app: FastAPI):
print('lifespan start')
yield
print('lifespan end')
# Pass the lifespan function to FastAPI
app = FastAPI(
docs_url=docs_url,
redoc_url=redoc_url,
openapi_url=openapi_url,
lifespan=lifespan
)
# Register routes
register_routes(app)
return app
# Instantiate the app
app = create_app()
...
def register_routes(app: FastAPI):
app.include_router(rabbit_router, include_in_schema=False)
```
|
closed
|
2024-12-15T08:31:39Z
|
2024-12-15T16:09:15Z
|
https://github.com/airtai/faststream/issues/1986
|
[
"bug"
] |
easyjoh
| 2
|
PokemonGoF/PokemonGo-Bot
|
automation
| 5,520
|
Can't get it to evolve (SOLVED CLOSE)
|
EDIT: FIGURED IT OUT CLOSE PLEASE, accidentally kept // in front of something that broke it
[2016-09-17 18:39:59] [MainThread] [EvolvePokemon] [WARNING] evolve_speed is deprecated, instead please use 'min_evolve_speed' and 'max_evolved_speed'.
However when I look at config https://github.com/PokemonGoF/PokemonGo-Bot/blob/master/docs/configuration_files.md#evolve-all-configuration it looks like this is correct notation
```
}, {
"type": "EvolvePokemon",
"config": {
"enabled": true,
"evolve_list": "pidgey, Nidoran F, Nidoran M, magnemite",
"donot_evolve_list": "none",
"first_evolve_by": "cp",
"evolve_above_cp": 100,
"evolve_above_iv": 0.8,
"logic": "or",
"min_evolve_speed": 16,
"max_evolve_speed": 20,
"use_lucky_egg": false
}
```
### Other Information
OS: Mac OSX
<!-- Tell us what Operating system you're using -->
Branch:
Git Commit: 11123a82
Python Version:
|
closed
|
2016-09-18T01:50:20Z
|
2016-09-18T04:57:44Z
|
https://github.com/PokemonGoF/PokemonGo-Bot/issues/5520
|
[] |
HelloTroy
| 2
|
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.