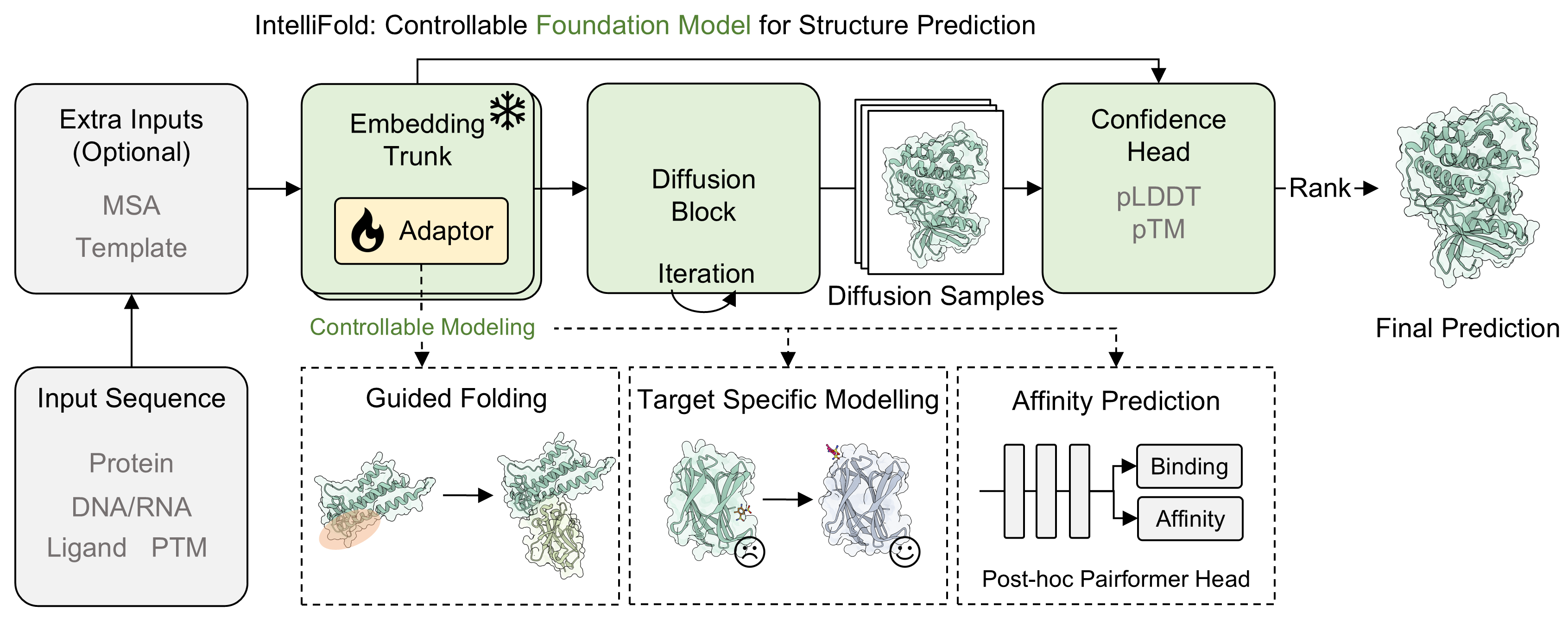
IntelliFold: A Controllable Foundation Model for General and Specialized Biomolecular Structure Prediction.
🎉 New Model Release
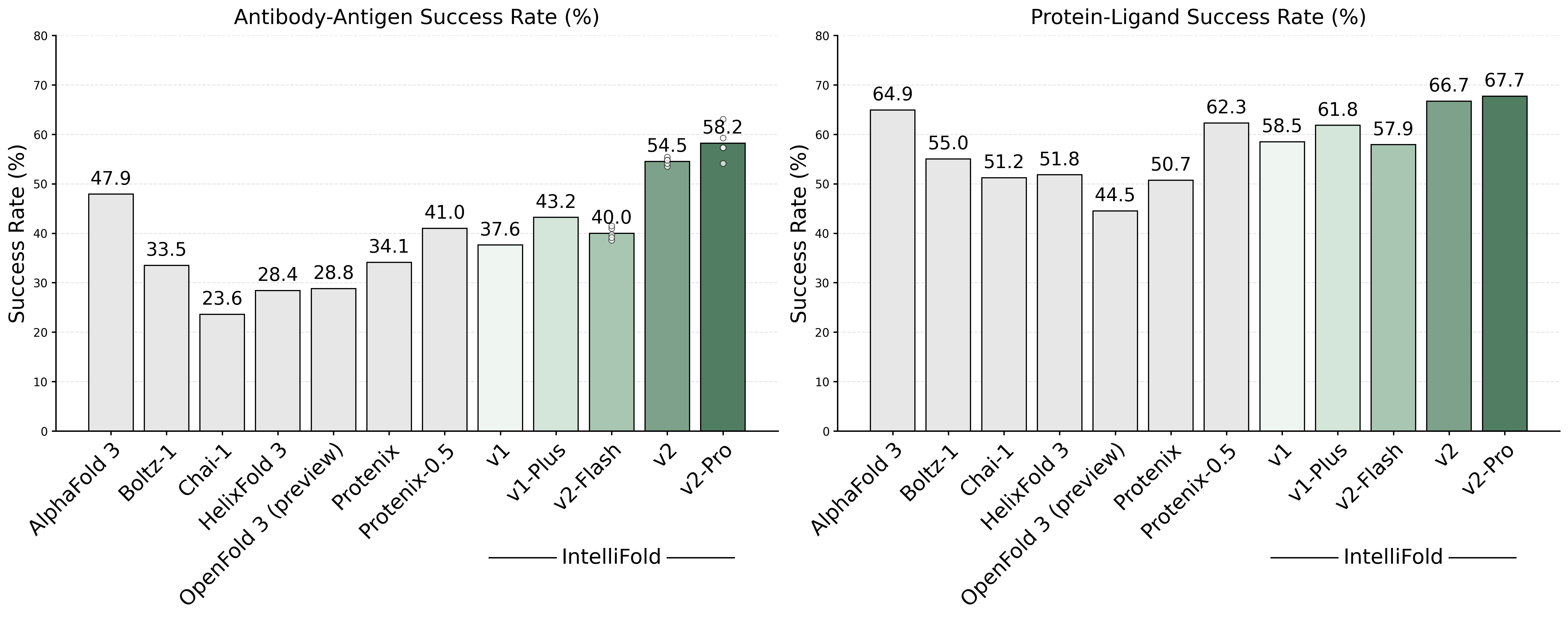
- 2026-02-07: We are excited to present [IntelliFold 2]. This version represents a major architectural update and is one of the first open-source models to outperform AlphaFold3 on Foldbench.
📊 Benchmarking
To comprehensively evaluate the performance of IntelliFold 2, we conducted a rigorous evaluation on FoldBench. We compared IntelliFold against several leading methods, including Boltz-1,2, Chai-1, Protenix and Alphafold3.
For more details on the benchmarking process and results, please refer to our release note IntelliFold 2 Release Note and IntelliFold Technical Report.
🚀 Quick Start
To quickly get started with IntelliFold, you can use the following commands:
# Install IntelliFold from PyPI
pip install intellifold
# Run inference with an example YAML file
intellifold predict ./examples/5S8I_A.yaml --out_dir ./output
⚙️ Installation
To more complete installation instructions and usage, please refer to the Installation Guide.
🔍 Inference
Prepare Input File: Create a YAML file with your sequences following our input format specification
Run Prediction:
intellifold predict your_input.yaml --out_dir ./resultsIntelliFold v2-Flash will be used by default, you can also use IntelliFold v2 by specifying the model name:
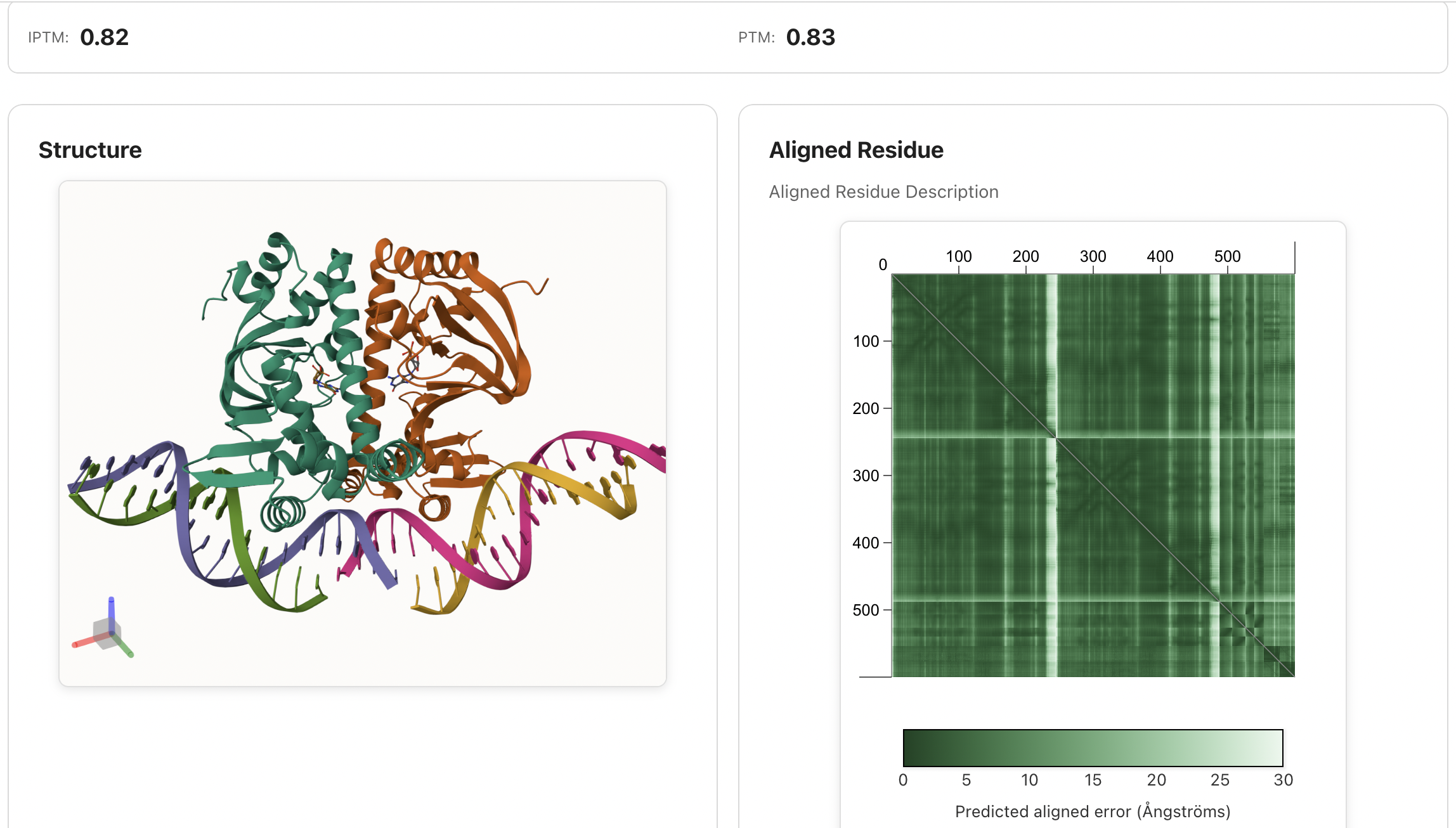
intellifold predict your_input.yaml --out_dir ./results --model v2Check Results: Find predicted structures and confidence scores in the output directory, you can also check the section of output format in output documentation.
Optional Optimization: Enable custom kernels for faster inference and reduced memory usage
For comprehensive usage instructions and examples, refer to the Usage Guide.
🌐 IntelliFold Server
We highly recommend using the IntelliFold Server for the most accurate, complete, and convenient biomolecular structure predictions. It requires no installation and provides an intuitive web interface to submit your sequences and visualize results directly in your browser. The server runs the full, optimized, latest IntelliFold implementation for optimal performance.
📜 Citation
If you use IntelliFold in your research, please cite our paper:
@techreport{qiao2026intellifold,
title={{IntelliFold 2: Surpassing AlphaFold 3 via Architectural Refinement and Structural Consistency}},
author={Lifeng Qiao and He Yan and Gary Liu and Gaoxing Guo and Siqi Sun},
year={2026},
institution={IntelliGen-AI},
type={Release Note},
url={https://raw.githubusercontent.com/IntelliGen-AI/IntelliFold/main/assets/Intellifold_v2_release_note.pdf}
}
@misc{theintfoldteam2025intfoldcontrollablefoundationmodel,
title={IntFold: A Controllable Foundation Model for General and Specialized Biomolecular Structure Prediction},
author={The IntFold Team and Leon Qiao and Wayne Bai and He Yan and Gary Liu and Nova Xi and Xiang Zhang},
year={2025},
eprint={2507.02025},
archivePrefix={arXiv},
primaryClass={q-bio.BM},
url={https://arxiv.org/abs/2507.02025}
}
🔗 Acknowledgements
- The implementation of fast layernorm operators is inspired by OneFlow and FastFold, following Protenix's usage.
- Many components in
intellifold/openfold/are adapted from OpenFold, with substantial modifications and improvements by our team (except for theLayerNormpart). - This repository, the implementation of Inference Data Pipeline(Data/Feature Processing and MSA generation tasks) referred to Boltz-1, and modify some codes to adapt to the input of our model.
⚖️ License
The IntelliFold project, including code and model parameters, is made available under the Apache 2.0 License, it is free for both academic research and commercial use.
📬 Contact Us
If you have any questions or are interested in collaboration, please feel free to contact us at contact@intfold.com.
- Downloads last month
- 629